
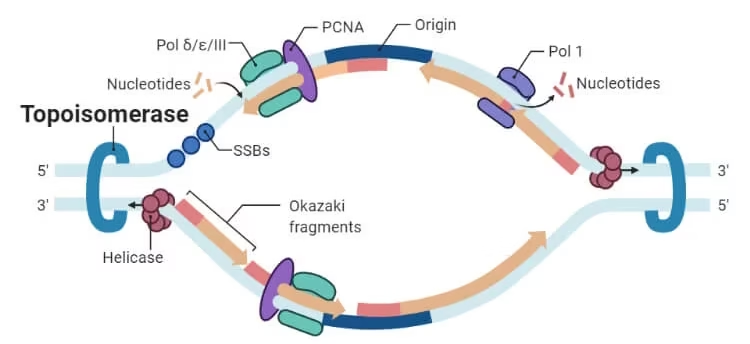
Topoisomerase
- DNA is not just a straight ladder — in the cell, it’s supercoiled, twisted, and packed tightly.
- Whenever DNA is opened (for replication, transcription, or repair), the double helix ahead of the opening gets overwound — like a phone cord twisting up.
- This creates torsional stress that can stall or damage the process.
Topoisomerases are DNA “untangling” enzymes that:
-
Temporarily cut one or both DNA strands.
-
Allow controlled movement of DNA to remove twists, knots, or tangles.
-
Reseal the DNA without leaving gaps or errors.
Without them:
-
DNA would become so twisted that replication forks and RNA polymerases could not move forward.
-
Chromosomes would remain knotted or linked after replication and could not be separated during cell division.
Differences:
-
Type I: Cuts one strand; works in smaller steps; mostly ATP-independent.
-
Type II: Cuts both strands; can pass large segments through each other; requires ATP.
Topoisomerase Types
Topoisomerases are classified in two main ways:
| Feature | Type I | Type II |
|---|---|---|
| Strands cut | One | Two |
| ATP use | Usually none (except reverse gyrase) | Yes |
| DNA movement | Single-strand rotation or passage | Double-strand passage |
| Functions | Relieve supercoils | Relieve supercoils, untangle DNA, introduce supercoils (in bacteria) |
Type-I Topoisomerase
Type I topoisomerases cut one DNA strand and allow the DNA to rotate or pass a strand through before resealing.
Think of it like untying a rope by loosening just one side.
Structure
-
Generally, single protein chains (monomers).
-
Active site tyrosine attacks the DNA backbone to form a temporary phosphotyrosyl bond.
-
Has a DNA-binding groove shaped to hold DNA in place during cutting.
-
Subfamilies differ in domain arrangement and how they move DNA.
Subtypes of Type I
-
Type IA
-
Cut one strand, pass the other strand through the gap.
-
Form 5′-phosphotyrosyl bonds.
-
Only relax negative supercoils.
-
Need single-stranded DNA regions to start.
-
Example: E. coli Topo I and Topo III.
-
-
Type IB
-
Cut one strand and let it freely rotate to release supercoiling.
-
Form 3′-phosphotyrosyl bonds.
-
Can relax both positive and negative supercoils.
-
Example: human Topo I.
-
-
Type IC
-
Found mostly in archaea (e.g., Topo V).
-
Mechanism like Type IB, but the structure is completely different.
-
Type IA Example: E. coli Topo I
-
Controls the negative supercoiling level in the cell.
-
Works with DNA gyrase to maintain proper DNA topology balance.
-
Topo III is more specialised for recombination intermediate resolution.
Mechanism of Type I Action
For Type IB (free rotation model):
-
The enzyme binds to the DNA helix.
-
Active site tyrosine attacks the phosphodiester backbone, forming a 3′-phosphotyrosyl bond.
-
The cut end of DNA swivels around the intact strand to relieve tension.
-
The enzyme uses the free –OH group on DNA to attack the phosphotyrosyl bond, sealing the break.
For Type IA (strand passage model):
-
Enzyme grips both single-stranded and double-stranded regions.
-
Cuts one strand and opens a gate.
-
Passes another strand through the gap.
-
Reseals the DNA.
Functions
-
Relieve negative supercoils during transcription/replication.
-
Resolve single-stranded knots.
-
Prepare DNA for packaging into nucleosomes (in eukaryotes).
-
Help in recombination and DNA repair.
Type-II Topoisomerase
Type II enzymes cut both strands of DNA at the same time, pass another double-stranded segment through, and reseal.
Think of it like undoing a knot in a rope by opening it completely and pulling another rope section through.
Structure
-
Usually multimeric:
-
Bacteria: heterotetramers (A₂B₂).
-
Eukaryotes: homodimers.
-
-
Three major regions:
-
ATPase domain (at the N-terminus) – binds/hydrolyses ATP to power conformational changes.
-
DNA-cleavage/religation core – contains the active site tyrosines (one per strand).
-
C-terminal domain – determines DNA preferences and cellular roles.
-
Subtypes
-
Type IIA: most common in bacteria and eukaryotes.
-
Examples: DNA gyrase, Topo IV, eukaryotic Topo IIα, Topo IIβ.
-
-
Type IIB: found in archaea and plants.
-
Example: Topo VI – related to the Spo11 protein in meiosis (causes programmed DSBs).
-
Mechanism of Type II Action
-
Bind the G-segment (the one to be cut) in the cleavage core.
-
Capture T-segment in the ATPase domain.
-
Hydrolyse ATP to close the ATPase gate, trapping the T-segment.
-
Cut both strands of G-segment, hold ends covalently via active site tyrosines.
-
Pass the T-segment through the break.
-
Reseal the G-segment and release the T-segment.
-
Hydrolyse ATP to reset the enzyme.
Functions
-
Decatenate (unlink) replicated chromosomes.
-
Relax both positive and negative supercoils.
-
In bacteria, gyrase introduces negative supercoils to compact DNA.
-
In eukaryotes, Topo IIα condenses chromosomes during mitosis.
Topoisomerase Inhibition
Topo I Inhibitors (mainly cancer drugs)
-
Camptothecin (natural alkaloid from Camptotheca acuminata).
-
Topotecan – used for ovarian and small-cell lung cancer.
-
Irinotecan – used for colorectal cancer.
-
Mechanism: stabilise the covalent Topo I–DNA complex → replication collision → DNA double-strand breaks.
Topo II Inhibitors (cancer drugs)
-
Etoposide and Teniposide – block resealing step.
-
Doxorubicin, Mitoxantrone – intercalate into DNA and trap Topo II.
-
Side effects: bone marrow suppression, possible secondary leukaemias (due to DNA damage in healthy cells).
Bacterial Topo II Inhibitors
-
Fluoroquinolones (ciprofloxacin, levofloxacin) – target gyrase and Topo IV.
-
Cause DNA double-strand breaks in bacteria → cell death.
-
Widely used but resistance is growing.
Clinical Significance
-
Essential for cell survival → attractive drug targets.
-
Cancer therapy: Topo poisons kill rapidly dividing cells.
-
Antibiotics: Target bacterial topoisomerases without affecting human ones.
-
Resistance mechanisms: mutations in binding sites, drug efflux, protective proteins.
Topoisomerase vs Helicase
-
Helicase: unwinds DNA strands using ATP.
-
Topoisomerase: relieves twisting pressure caused by helicase and other processes.
-
Without topoisomerase, helicase would stall due to supercoiling ahead of the fork.
| Feature | Topoisomerase | Helicase |
|---|---|---|
| Main job | Removes supercoils, untangles DNA, and separates linked DNA circles | Unwinds the double helix into two single strands |
| Action | Cuts DNA (one or both strands), moves DNA to relieve twist, then reseals | Breaks hydrogen bonds between bases to separate strands |
| DNA strands cut? | Yes – Type I cuts one, Type II cuts both | No cutting – just strand separation |
| ATP usage | Type I: usually no (except reverse gyrase); Type II: yes | Yes – requires ATP hydrolysis to move along DNA |
| When it works | Ahead of replication forks, transcription bubbles, and during chromosome segregation | At replication forks during DNA synthesis |
| Directionality | Not strongly directional – acts where needed | Moves in a specific 5′→3′ or 3′→5′ direction along DNA |
| Effect on DNA | Changes DNA topology (supercoiling, knotting, catenation) | Produces single-stranded templates for replication or repair |
| Coordination | Works with helicase – prevents overwinding caused by strand separation | Works with topoisomerase – creates the supercoils that need to be relieved |
Topoisomerase vs Gyrase
-
Gyrase is a bacterial Type II topoisomerase that can introduce negative supercoils.
-
Helps bacteria compact DNA and make it ready for transcription.
-
Targeted by antibiotics like fluoroquinolones.
-
No equivalent enzyme in humans → good selective drug target.
| Feature | Topoisomerase (General) | DNA Gyrase |
|---|---|---|
| Definition | Enzymes that change DNA topology by cutting and rejoining DNA strands to remove or add supercoils | A Type II bacterial topoisomerase that can introduce negative supercoils into DNA |
| Occurrence | Found in all organisms (bacteria, archaea, eukaryotes) | Found only in bacteria and some archaea |
| Types | Type I (cuts 1 strand) and Type II (cuts 2 strands) | Type II only (heterotetramer: GyrA₂GyrB₂) |
| Supercoil activity | Mostly relaxes supercoils; some (like reverse gyrase) add positive supercoils | Introduces negative supercoils and can also relax supercoils |
| ATP requirement | Type I: usually no; Type II: yes | Yes – requires ATP hydrolysis |
| Functions | Relieve torsional stress, untangle DNA, separate linked chromosomes | Maintain negative supercoiling in bacterial chromosomes; assist in replication, transcription, recombination |
| Drug targets | Anti-cancer (e.g., etoposide, camptothecin) and antibacterial (fluoroquinolones target bacterial topoisomerases) | Primary target of many fluoroquinolone antibiotics (e.g., ciprofloxacin) |
| Presence in humans | Yes – humans have several topoisomerases | No – gyrase is absent in humans, making it a good selective antibacterial target |