
Introduction
- Nucleic acids are biological macromolecules responsible for storing and transmitting genetic information.
- They exist in all living organisms and viruses.
- The two main types are:
-
DNA (Deoxyribonucleic Acid): Double-stranded molecule that serves as the genetic blueprint.
-
RNA (Ribonucleic Acid): Usually single-stranded, involved in gene expression and regulation.
-
- Studying nucleic acids requires their isolation in pure, intact, and concentrated form.
- Crude extracts from cells contain proteins, polysaccharides, lipids, pigments, and secondary metabolites that interfere with molecular techniques.
- Therefore, specialized methods have been developed to extract and purify DNA and RNA.
Importance of Nucleic Acid Isolation
-
Genetic Studies – Understanding hereditary traits, mutations, and genome organization.
-
Medical Applications – Diagnosis of infectious diseases (PCR for TB, HIV, COVID-19, etc.).
-
Forensic Science – DNA fingerprinting for identification, paternity testing, and criminal investigations.
-
Agricultural Biotechnology – Genetic modification of crops, marker-assisted breeding.
-
Pharmaceutical Research – Development of vaccines, recombinant proteins, and therapeutic RNAs.
-
Evolutionary Studies – Comparative genomics, phylogenetics, mitochondrial DNA analysis.
Without efficient nucleic acid isolation, these fields would not be possible.
General Principles of Isolation
Although the exact procedure varies depending on the type of nucleic acid and source material, the basic principles are:
-
Cell Disruption (Lysis):
-
Breaking the cell membrane and/or cell wall to release contents.
-
Methods: grinding, enzymatic digestion, detergents, sonication, or freezing-thawing.
-
-
Separation of Nucleic Acids from Other Biomolecules:
-
Proteins removed using proteases or phenol extraction.
-
Carbohydrates removed with detergents or CTAB.
-
Lipids removed with organic solvents.
-
-
Selective Isolation:
-
Use of RNase to remove RNA during DNA extraction.
-
Use of DNase to remove DNA during RNA extraction.
-
-
Purification:
-
Precipitation with alcohol (ethanol or isopropanol).
-
Column chromatography (silica membranes).
-
Magnetic beads for automated purification.
-
-
Storage:
-
DNA stable at –20 °C or –80 °C.
-
RNA requires –80 °C with RNase inhibitors.
-
Types of Nucleic Acids Extracted
-
Genomic DNA (gDNA): Chromosomal DNA of an organism.
-
Plasmid DNA: Extra-chromosomal, circular DNA in bacteria.
-
Organelle DNA: Mitochondrial and chloroplast DNA.
-
Total RNA: Includes mRNA, tRNA, rRNA, and small RNAs.
-
mRNA: Messenger RNA used in gene expression analysis.
-
Small RNAs: Regulatory RNAs like miRNA, siRNA.
Methods of DNA Isolation
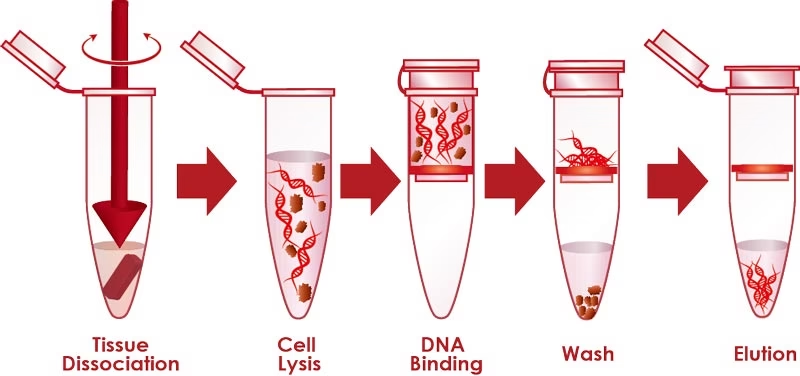 A. Genomic DNA Isolation
A. Genomic DNA Isolation
Example Sources: Animal tissues, plant leaves, bacterial cultures, blood samples.
Steps:
-
Sample Preparation: Fresh or frozen tissue ground in buffer.
-
Cell Lysis:
-
Animal cells: detergents like SDS or Triton X-100.
-
Plant cells: liquid nitrogen + CTAB buffer.
-
Bacteria: lysozyme digestion of peptidoglycan wall.
-
-
Protein Removal:
-
Proteinase K digests proteins.
-
Phenol-chloroform extraction separates proteins into organic phase.
-
-
DNA Precipitation:
-
DNA is precipitated with ethanol or isopropanol in presence of NaCl.
-
DNA appears as white fibrous threads.
-
-
Purification and Storage:
-
Washed with 70% ethanol.
-
Dissolved in TE buffer (Tris-EDTA).
-
Applications: Genome sequencing, PCR, cloning, gene mapping, diagnostics.
B. Plasmid DNA Isolation
Key Method: Alkaline Lysis
Steps:
-
Grow bacteria with plasmid on selective media (antibiotic resistance marker).
-
Lyse cells with alkaline SDS (denatures DNA and proteins).
-
Neutralize with potassium acetate – plasmid DNA renatures, chromosomal DNA precipitates.
-
Centrifuge and purify plasmid DNA.
-
Use column kits or ethanol precipitation for clean preparation.
Applications: Cloning, gene therapy vectors, recombinant protein expression.
C. Mitochondrial and Chloroplast DNA
-
Isolated using differential centrifugation to separate organelles.
-
Lysis of organelles followed by standard DNA extraction methods.
-
Useful for evolutionary biology, maternal lineage studies, and metabolic research.
Methods of RNA Isolation
RNA is more challenging than DNA because it is unstable and prone to RNase degradation.
Common Methods:
-
TRIzol Method (Phenol-Guanidinium):
-
Lysis buffer contains guanidinium isothiocyanate (denatures proteins and inactivates RNases).
-
After chloroform addition, solution separates into phases:
-
Aqueous phase (RNA)
-
Interphase (DNA)
-
Organic phase (proteins, lipids).
-
-
RNA is precipitated from aqueous phase with isopropanol.
-
-
Column-Based RNA Isolation:
-
Silica membrane binds RNA in presence of chaotropic salts.
-
After washing, pure RNA is eluted.
-
Fast, reproducible, widely used in labs.
-
-
Magnetic Bead-Based Method:
-
RNA binds to coated magnetic beads.
-
Automated platforms enable high-throughput RNA extraction.
-
Precautions:
-
Use RNase-free plasticware.
-
Wear gloves to prevent RNase contamination.
-
Use DEPC-treated water for RNA handling.
Applications: cDNA synthesis, RT-PCR, RNA sequencing, transcriptomics, gene expression analysis.
Specialized Isolation Methods
A. CTAB Method (Plants)
-
Plants contain polysaccharides and polyphenols that interfere with DNA.
-
CTAB (cationic detergent) forms insoluble complexes with polysaccharides, leaving DNA in solution.
B. Blood DNA Extraction
-
RBCs lack nuclei, so DNA is isolated from WBCs.
-
Lysis buffer, protease treatment, and precipitation yield clean genomic DNA.
C. Viral Nucleic Acid Extraction
-
Important for viral diagnostics (COVID-19, HIV, Hepatitis).
-
Kits optimized to isolate viral RNA/DNA from blood, swabs, or serum.
Quality and Quantity Assessment
A. Spectrophotometry
-
DNA and RNA absorb at 260 nm.
-
A260/A280 ratio:
-
Pure DNA ~1.8
-
Pure RNA ~2.0
-
-
A260/A230 ratio: >2 indicates minimal contamination.
B. Gel Electrophoresis
-
DNA: single high molecular weight band.
-
RNA: distinct 28S and 18S rRNA bands in eukaryotes.
C. Fluorescent Dyes
-
PicoGreen for DNA, RiboGreen for RNA.
-
Very sensitive, useful for low concentration samples.
Challenges in Isolation
-
Degradation by Nucleases
-
RNases are very stable and resistant to autoclaving.
-
Strict RNase-free techniques are essential.
-
-
Contamination
-
Protein contamination lowers purity.
-
Phenolic carryover interferes with PCR.
-
-
Sample Quality
-
Old or degraded tissue gives poor yields.
-
-
High Cost of Kits
-
Phenol-based methods are cheaper but hazardous.
-
Kits are safer but expensive.
-
Recent Advances
-
Automated Extraction Systems
-
Robots that process dozens of samples at once.
-
Used in hospitals for high-throughput diagnostics.
-
-
Microfluidic Devices
-
Portable chip-based systems for point-of-care testing.
-
-
Magnetic Nanoparticles
-
High efficiency, faster isolation, easy automation.
-
-
Single-Cell Nucleic Acid Isolation
-
Allows transcriptomics at single-cell resolution.
-
Important for cancer biology and developmental studies.
-
Applications
-
PCR and qPCR – Amplification of DNA and RNA.
-
Gene Cloning – Insertion of DNA into plasmids.
-
Sequencing – Next-generation sequencing requires high-quality DNA/RNA.
-
Diagnostics – Viral load detection, genetic disease screening.
-
Forensic Studies – DNA fingerprinting, identity verification.
-
Functional Genomics – Gene expression studies using mRNA and small RNAs.