Introduction
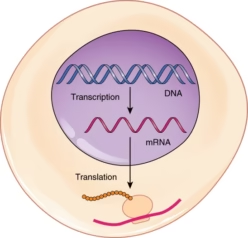
The flow of genetic information in all living organisms follows the central dogma of molecular biology, which states that DNA → RNA → Protein.
-
Transcription is the process of copying genetic information from DNA into RNA.
-
Translation is the process of converting the RNA sequence into a specific protein.
-
These processes ensure that genetic information stored in DNA is expressed as functional proteins, which carry out structural and metabolic roles in the cell.
-
Regulation of transcription and translation is essential for proper growth, development, and adaptation of organisms.
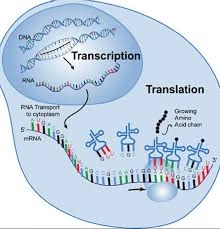
Factors Involved in Transcription and Translation
In Transcription
-
DNA Template: Provides the information for RNA synthesis.
-
RNA Polymerase: Enzyme that synthesizes RNA from DNA template.
-
Promoter Region: DNA sequence where RNA polymerase binds to initiate transcription.
-
Transcription Factors (in eukaryotes): Proteins that help RNA polymerase recognize promoter and regulate gene expression.
-
Ribonucleotides (ATP, GTP, UTP, CTP): Building blocks of RNA.
-
Regulatory Sequences: Enhancers, silencers, and operators that control transcription.
In Translation
-
mRNA: Carries the genetic code from DNA to ribosome.
-
Ribosome: Site of protein synthesis, made of rRNA and proteins.
-
tRNA: Brings amino acids to ribosome according to codon sequence.
-
Amino Acids: Building blocks of proteins.
-
Enzymes (Aminoacyl-tRNA synthetase): Attach correct amino acid to its tRNA.
-
Initiation, Elongation, and Release Factors: Proteins that control different stages of translation.
-
GTP/ATP: Energy sources for translation process.
RNA Processing
- In eukaryotic cells, the RNA formed directly after transcription is called the primary transcript (pre-mRNA).
- It is not functional and undergoes several modifications to become mature mRNA that can be translated into a protein.

1. Capping (5’ Cap Formation)
-
A 7-methylguanosine (m7G) cap is added to the 5’ end of the pre-mRNA.
-
Functions:
-
Protects mRNA from degradation by exonucleases.
-
Helps in ribosome recognition and initiation of translation.
-
2. Polyadenylation (Poly-A Tail Addition)
-
At the 3’ end, a chain of adenine nucleotides (Poly-A tail) is added by poly(A) polymerase.
-
Functions:
-
Increases stability of mRNA.
-
Facilitates transport of mRNA from nucleus to cytoplasm.
-
Helps in efficient translation.
-
3. Splicing
-
Eukaryotic genes contain introns (non-coding regions) and exons (coding regions).
-
Splicing removes introns and joins exons to form a continuous coding sequence.
-
Carried out by a complex called spliceosome (made of snRNA + proteins).
-
Alternative splicing: A single gene can give rise to different proteins by joining exons in different combinations.
4. RNA Editing (in some cases)
-
Nucleotides of RNA may be inserted, deleted, or chemically modified.
-
Changes the coding information.
-
Example: ApoB100 → ApoB48 editing in humans.
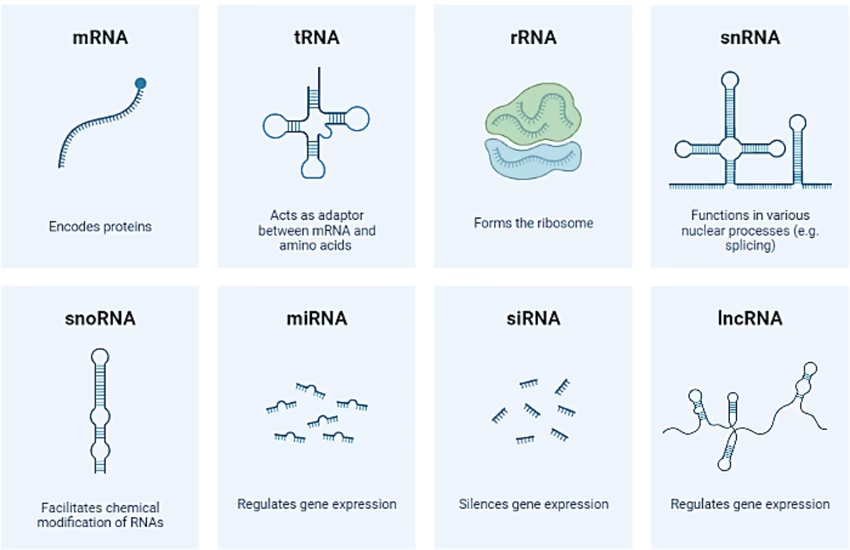
Types of RNA
-
mRNA (Messenger RNA): Carries genetic code from DNA to ribosomes.
-
tRNA (Transfer RNA): Brings specific amino acids during translation.
-
rRNA (Ribosomal RNA): Structural and catalytic component of ribosomes.
-
snRNA (Small nuclear RNA): Involved in splicing of pre-mRNA.
-
miRNA (Micro RNA) & siRNA (Small interfering RNA): Regulate gene expression by silencing or degrading mRNA.
-
lncRNA (Long non-coding RNA): Regulatory functions in gene expression.
Genetic Code
- The genetic code is the set of rules by which the sequence of nucleotides in mRNA is translated into the sequence of amino acids in a protein.
- It is universal for almost all organisms and ensures that the genetic information in DNA is correctly expressed as proteins.
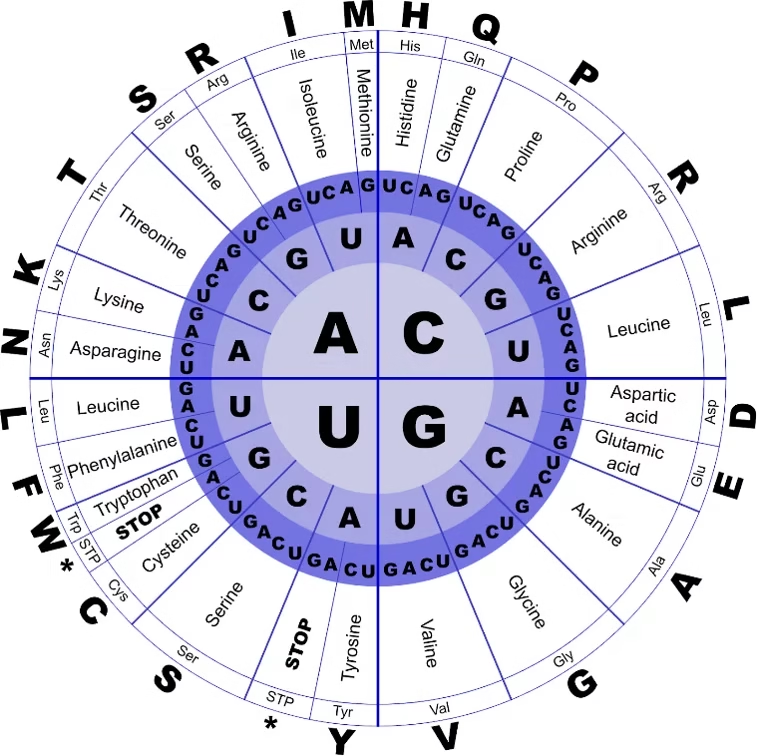
Features of the Genetic Code
-
Triplet Code
-
Each amino acid is encoded by a sequence of three nucleotides (codon).
-
Example: AUG codes for Methionine.
-
-
Universal
-
The same codon codes for the same amino acid in almost all living organisms (bacteria to humans).
-
-
Degenerate (Redundant)
-
Most amino acids are coded by more than one codon.
-
Example: Leucine is coded by six codons (UUA, UUG, CUU, CUC, CUA, CUG).
-
-
Unambiguous
-
Each codon specifies only one amino acid.
-
-
Start Codon
-
AUG is the start codon and also codes for Methionine (initiates translation).
-
-
Stop Codons
-
UAA, UAG, UGA → Do not code for any amino acid.
-
They signal termination of protein synthesis.
-
-
Non-overlapping and Commaless
-
Codons are read one after another, without overlapping or punctuation.
-
Types of Codons
-
Sense codons: 61 codons that specify amino acids.
-
Nonsense codons: 3 stop codons (UAA, UAG, UGA).
Lac Operon
- The lac operon is a gene regulatory system in Escherichia coli (E. coli) that controls the metabolism of lactose.
- It is a classic example of an inducible operon, meaning it is usually OFF but can be switched ON in the presence of lactose.
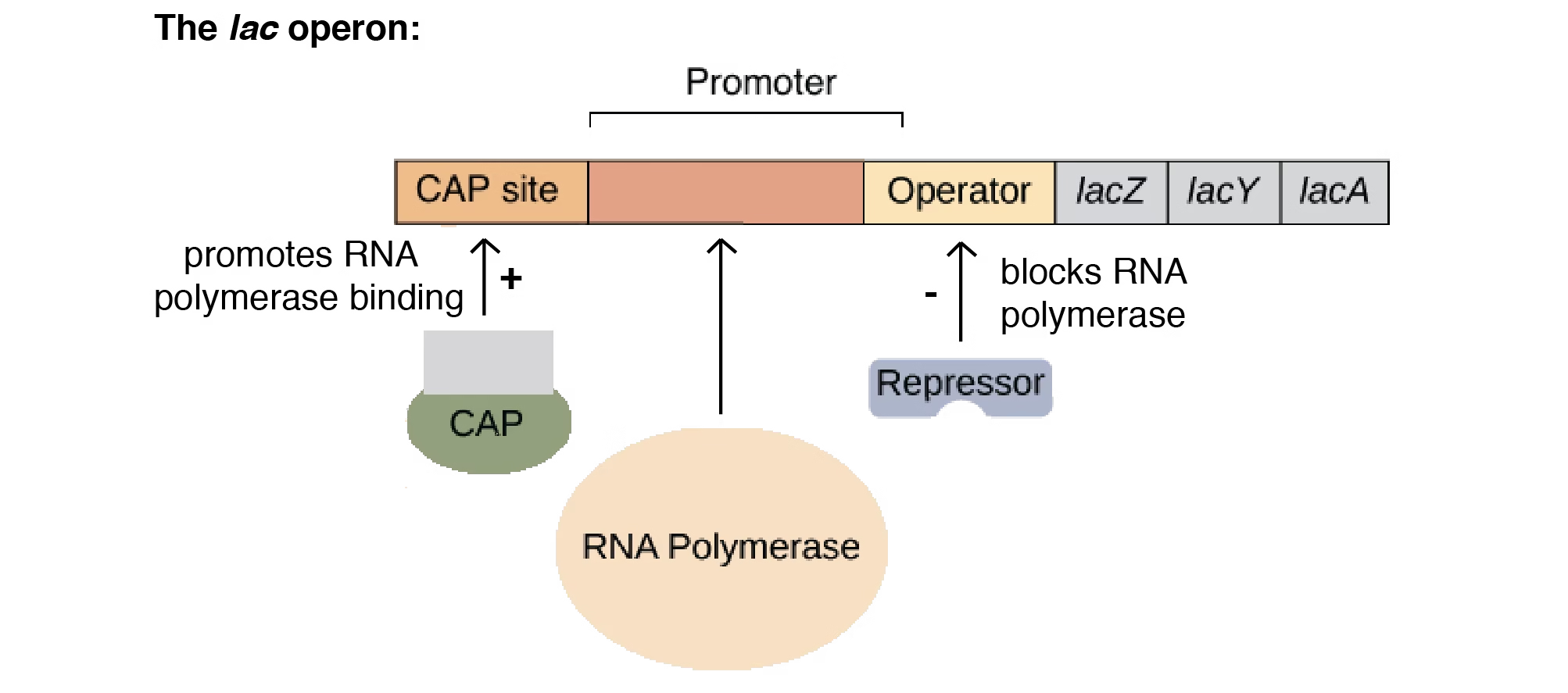
Components of Lac Operon
-
Structural Genes
-
lacZ → codes for β-galactosidase (breaks lactose into glucose + galactose).
-
lacY → codes for permease (transports lactose into the cell).
-
lacA → codes for transacetylase (detoxifies by-products).
-
-
Regulatory Elements
-
Promoter (P): Site where RNA polymerase binds to start transcription.
-
Operator (O): Site where the repressor binds to block transcription.
-
Regulator Gene (lacI): Produces repressor protein that controls the operon.
-
Working of Lac Operon
-
Without Lactose (Operon OFF):
-
Repressor protein binds to operator.
-
RNA polymerase cannot move forward.
-
No transcription of structural genes.
-
-
With Lactose (Operon ON):
-
Lactose is converted into allolactose (inducer).
-
Allolactose binds to repressor → inactivates it.
-
RNA polymerase binds promoter and transcribes structural genes.
-
Enzymes are produced for lactose metabolism.
-
Regulation by Glucose (Catabolite Repression)
-
When glucose is present, the lac operon remains mostly OFF even if lactose is available.
-
cAMP-CAP complex is required for efficient transcription.
-
Low glucose → High cAMP → cAMP binds CAP → enhances transcription.
-
High glucose → Low cAMP → CAP does not bind → transcription reduced.
-
Tryptophan (Trp) Operon
- The tryptophan operon is a gene regulatory system in Escherichia coli (E. coli) that controls the biosynthesis of the amino acid tryptophan.
- It is a classic example of a repressible operon, meaning it is usually ON but can be turned OFF when tryptophan is abundant.
Components of Trp Operon
-
Structural Genes (trpE, trpD, trpC, trpB, trpA)
-
Encode enzymes required for synthesis of tryptophan.
-
-
Regulatory Elements
-
Promoter (P): Site where RNA polymerase binds.
-
Operator (O): Binding site for the repressor.
-
Regulator Gene (trpR): Produces an inactive repressor protein.
-
Leader Sequence (trpL): Involved in fine regulation by attenuation.
-
Mechanism of Regulation
-
When Tryptophan is Absent (Operon ON):
-
Repressor protein is inactive and cannot bind the operator.
-
RNA polymerase binds to promoter and transcribes the structural genes.
-
Enzymes are synthesized → Tryptophan is produced.
-
-
When Tryptophan is Present (Operon OFF):
-
Tryptophan acts as a co-repressor.
-
It binds to the inactive repressor protein, activating it.
-
The active repressor binds to the operator and blocks RNA polymerase.
-
Transcription of structural genes stops → No unnecessary tryptophan synthesis.
-
Attenuation (Extra Control Mechanism)
-
In addition to repression, the trp operon also uses attenuation for regulation.
-
The leader region (trpL) contains short sequences that can form hairpin loops in mRNA.
-
When tryptophan levels are high → ribosome moves quickly → forms a terminator loop → transcription stops early.
-
When tryptophan levels are low → ribosome stalls → forms anti-terminator loop → transcription continues.
Regulation in Eukaryotes
Gene regulation in eukaryotes is more complex than in prokaryotes because:
-
DNA is packaged into chromatin.
-
Genes are separated by introns and exons.
-
Multiple levels of control exist (before transcription to after protein synthesis).
This regulation ensures cell differentiation, development, and adaptation.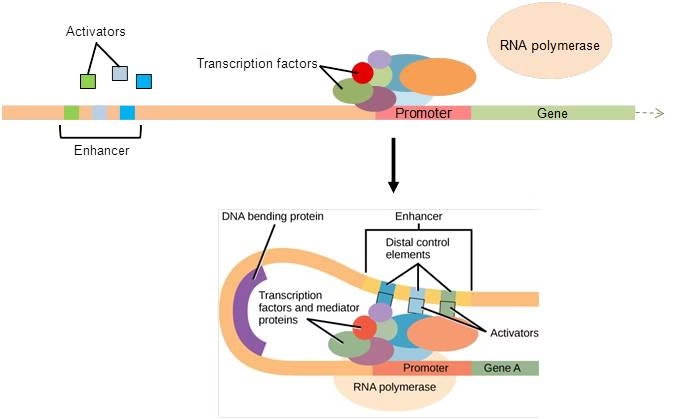
Levels of Gene Regulation in Eukaryotes
1. Epigenetic Regulation (Chromatin Level)
-
Histone Modification: Acetylation, methylation, phosphorylation of histones control accessibility of DNA.
-
Histone acetylation: Opens chromatin → increases transcription.
-
Histone deacetylation: Condenses chromatin → decreases transcription.
-
-
DNA Methylation: Addition of methyl groups to cytosine → silences genes.
-
Chromatin Remodeling Complexes: Rearrange nucleosomes to allow or block transcription.
2. Transcriptional Regulation
-
Promoters: DNA sequences where RNA polymerase binds.
-
Enhancers & Silencers: Regulatory sequences that increase or decrease transcription.
-
Transcription Factors: Proteins that bind DNA to regulate gene expression (activators & repressors).
-
Mediator Complex: Connects transcription factors with RNA polymerase.
3. Post-Transcriptional Regulation
-
Alternative Splicing: A single pre-mRNA can give rise to different mRNAs and proteins.
-
RNA Editing: Nucleotide modifications can change coding information.
-
RNA Transport: Export of mRNA from nucleus to cytoplasm can be controlled.
-
mRNA Stability: Poly-A tail length and binding proteins regulate how long mRNA remains functional.
4. Translational Regulation
-
Control of Initiation: Proteins and initiation factors regulate the binding of ribosomes to mRNA.
-
miRNA & siRNA: Small RNAs bind to mRNA and prevent translation or degrade it (RNA interference).
5. Post-Translational Regulation
-
Protein Folding & Modifications: Proteins may undergo phosphorylation, glycosylation, acetylation.
-
Protein Degradation: Ubiquitin-proteasome system marks proteins for destruction.
-
Compartmentalization: Proteins activated only when transported to correct organelle.
Gene Dosage and Gene Amplification
1. Gene Dosage
-
Definition: Gene dosage refers to the number of copies of a particular gene present in a cell or organism.
-
Normally, each gene is present in two copies (diploid) in humans.
-
If the number of gene copies changes, the expression level of that gene also changes.
Effects of Gene Dosage:
-
Increased dosage: More copies → excess production of gene product (protein).
-
Decreased dosage: Fewer copies → reduced gene product.
Examples:
-
Down Syndrome (Trisomy 21): Extra copy of chromosome 21 → higher gene dosage.
-
Turner Syndrome (XO): Only one X chromosome → lower gene dosage of X-linked genes.
2. Gene Amplification
-
Definition: Gene amplification is the increase in the number of copies of a specific gene within a cell.
-
It is a controlled process and often occurs when cells need large amounts of a specific product.
Examples:
-
rRNA Genes: Amplified in rapidly dividing cells to meet high protein synthesis demand.
-
Oncogenes in Cancer: Amplification of genes like HER2/neu in breast cancer leads to uncontrolled cell growth.
-
In Insects: Amplification of detoxifying enzyme genes provides resistance against pesticides.
Difference Between Gene Dosage and Gene Amplification
| Feature | Gene Dosage | Gene Amplification |
|---|---|---|
| Definition | Change in number of whole gene copies due to chromosomal abnormalities. | Increase in copies of a particular gene within the genome. |
| Cause | Chromosome gain/loss (aneuploidy). | Specific gene replication. |
| Effect | Global effect on all genes of that chromosome. | Local effect on a single/few genes. |
| Example | Down syndrome (extra chromosome 21). | HER2 gene amplification in cancer. |
Generation of Antibody Diversity
- The immune system can produce millions of different antibodies to recognize a vast array of antigens, even though the genome contains a limited number of antibody genes.
- This diversity is generated by several mechanisms in B-cells.
1. V(D)J Recombination
-
Definition: Random rearrangement of gene segments in B-cells.
-
Components:
-
V (Variable) segments
-
D (Diversity) segments – only in heavy chains
-
J (Joining) segments
-
-
During B-cell development, different V, D, and J segments are combined to create a unique variable region of the antibody.
-
Result: Generates a large variety of antigen-binding sites.
2. Junctional Diversity
-
During V(D)J recombination, random addition or deletion of nucleotides occurs at the joining sites.
-
This further increases variability in the antibody’s antigen-binding site.
3. Somatic Hypermutation
-
Occurs after antigen stimulation in mature B-cells.
-
Introduces point mutations in the variable region of antibody genes.
-
B-cells producing higher-affinity antibodies are selected → affinity maturation.
4. Class Switch Recombination (Isotype Switching)
-
Changes the constant region of the antibody heavy chain without altering the antigen specificity.
-
Allows the antibody to switch from IgM → IgG, IgA, or IgE depending on the immune response.
5. Combinatorial Association
-
Each antibody has two heavy chains and two light chains.
-
Different combinations of heavy and light chains further increase diversity.