
Introduction
-
Mutation refers to a heritable and permanent change in the nucleotide sequence of genetic material (DNA or RNA).
-
Mutations are fundamental biological events that generate genetic variation.
-
Genetic variability produced by mutations is essential for evolution, adaptation, and natural selection.
-
Mutations can disrupt normal cellular functions and may result in:
-
Genetic disorders
-
Cancer
-
Congenital abnormalities
-
Microbial drug resistance
-
-
Mutations may occur spontaneously due to intrinsic cellular processes such as DNA replication errors or spontaneous base changes.
-
Mutations can also be induced by exposure to physical and chemical agents, known as mutagens.
-
In molecular biology and biotechnology, controlled mutagenesis is widely used to:
-
Study gene function
-
Analyze protein structure–function relationships
-
Improve industrial microbial strains
-
Types of Mutations
 1. Based on Origin
1. Based on Origin
-
Spontaneous mutations
-
Occur naturally without external agents
-
-
Induced mutations
-
Caused by physical or chemical mutagens
-
2. Based on Molecular Change
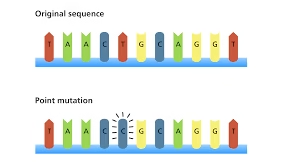
a. Point Mutations (Base Substitution)
-
Silent mutation – no change in amino acid
-
Missense mutation – change in amino acid
-
Nonsense mutation – premature stop codon
b. Frame Shift Mutations
-
Caused by insertion or deletion of nucleotides
-
Alters reading frame
-
Usually severe
3. Based on Effect on Protein
-
Silent mutations – no phenotypic effect
-
Loss-of-function mutations
-
Gain-of-function mutations
4. Based on Direction of Mutation
-
Forward mutation – wild type → mutant
-
Reverse mutation (reversion) – mutant → wild type
5. Based on Cell Type Affected
-
Somatic mutations
-
Occur in body cells
-
Not inherited
-
-
Germ line mutations
-
Occur in reproductive cells
-
Inherited
-
6. Based on Effect on Viability
-
Lethal mutations
-
Sub-lethal mutations
-
Neutral mutations
7. Based on Nutritional Requirement (Microbial Genetics)
-
Auxotrophic mutations
-
Prototrophic (wild type)
8. Based on Conditional Expression
-
Conditional mutations
-
Expressed only under specific conditions
-
Example: temperature-sensitive mutants
-
Spontaneous Mutation
A spontaneous mutation is a heritable change in the genetic material that occurs naturally without exposure to any external mutagen. These mutations arise from normal cellular processes and the inherent chemical instability of DNA.
Causes of Spontaneous Mutation
1. Errors During DNA Replication
-
Incorrect nucleotide incorporation by DNA polymerase
-
Failure of proofreading (3′→5′ exonuclease activity)
-
Leads mainly to base substitution mutations
2. Tautomeric Shifts
-
Nitrogenous bases exist in rare tautomeric forms
-
Abnormal base pairing occurs:
-
Tautomeric adenine pairs with cytosine
-
Tautomeric thymine pairs with guanine
-
-
Results in transition mutations
3. Depurination
-
Loss of purine bases (adenine or guanine) from DNA
-
Produces apurinic (AP) sites
-
During replication, incorrect bases may be inserted
4. Deamination
-
Removal of amino groups from bases:
-
Cytosine → Uracil
-
Adenine → Hypoxanthine
-
Guanine → Xanthine
-
-
Causes transition mutations if unrepaired
5. Oxidative Damage
-
Reactive oxygen species (ROS) damage DNA bases
-
Example: formation of 8-oxoguanine
-
Leads to mispairing and mutations
6. Transposable Elements
-
“Jumping genes” move within the genome
-
Cause insertions, deletions, or gene disruption
Characteristics of Spontaneous Mutations
-
Occur at a low but constant rate
-
Random and non-directional
-
Usually corrected by DNA repair mechanisms
-
Major source of natural genetic variation
Biological Significance
-
Provide raw material for evolution
-
Contribute to genetic diversity
-
Influence adaptation and survival
Medical Significance
-
Cause of:
-
Genetic disorders
-
Cancer (somatic mutations)
-
-
Germ-line mutations are heritable
Induced Mutation
An induced mutation is a heritable change in the genetic material that occurs due to exposure to external agents known as mutagens. Unlike spontaneous mutations, induced mutations occur at a much higher frequency.
Mutagens Responsible for Induced Mutation
1. Physical Mutagens
-
Ultraviolet (UV) radiation – causes thymine dimers
-
Ionizing radiation (X-rays, gamma rays) – causes DNA strand breaks
2. Chemical Mutagens
-
Base analogs (e.g., 5-bromouracil)
-
Deaminating agents (e.g., nitrous acid)
-
Alkylating agents (e.g., EMS)
-
Intercalating agents (e.g., acridine dyes)
3. Biological Mutagens
-
Viruses
-
Transposable elements (jumping genes)
Mechanism of Induced Mutation
-
Mutagens interact with DNA causing:
-
Base substitutions
-
Insertions or deletions
-
DNA strand breaks
-
-
If DNA repair mechanisms fail, mutations become permanent
Characteristics of Induced Mutations
-
Occur at high frequency
-
Can be controlled experimentally
-
Often dose-dependent
-
May be localized or random
Types of Induced Mutations
-
Point mutations
-
Frame shift mutations
-
Chromosomal mutations
Applications of Induced Mutation
Research Applications
-
Studying gene function
-
Mapping metabolic pathways
Industrial Applications
-
Strain improvement for:
-
Antibiotic production
-
Enzyme synthesis
-
Vitamin production
-
Medical Importance
-
Role in:
-
Cancer development
-
Genetic disorders
-
-
Basis for toxicity and carcinogenicity testing (Ames test)
Silent Mutation
A silent mutation is a type of point mutation in which a change in the nucleotide sequence of DNA does not result in a change in the amino acid sequence of the protein.
Why It Is Called “Silent”
-
The genetic code is degenerate, meaning:
-
More than one codon can code for the same amino acid
-
-
Even after a base change, the same amino acid is incorporated
-
Hence, no obvious change in protein structure or function occurs
Molecular Basis of Silent Mutation
-
Occurs due to base substitution (usually transition)
-
The altered codon still codes for the same amino acid
-
Commonly seen at the third position of codon (wobble position)
Example
-
Original codon: GAA → codes for Glutamic acid
-
Mutated codon: GAG → also codes for Glutamic acid
➡ Amino acid sequence remains unchanged
Effects of Silent Mutation
-
Usually no phenotypic effect
-
Protein structure and function remain normal
However, in some cases silent mutations may:
-
Affect mRNA stability
-
Alter translation efficiency
-
Influence protein folding indirectly
Detection of Silent Mutations
-
Cannot be detected by protein analysis
-
Detected by:
-
DNA sequencing
-
Molecular genetic techniques
-
Significance of Silent Mutations
Biological Significance
-
Contribute to genetic variation
-
Used as molecular markers in population genetics
Medical Significance
-
Important in:
-
Pharmacogenomics
-
Disease association studies
-
-
May affect gene expression levels
Comparison: Silent vs Missense Mutation
| Feature | Silent Mutation | Missense Mutation |
|---|---|---|
| Amino acid change | No | Yes |
| Protein function | Unchanged | Altered |
| Severity | Usually harmless | Variable |
Frame Shift Mutation
A frame shift mutation is a type of gene mutation caused by the insertion or deletion (indel) of one or more nucleotides in a DNA sequence not in multiples of three, resulting in a shift of the reading frame during translation.
Why It Is Called “Frame Shift”
-
The genetic code is read in triplets (codons)
-
Insertion or deletion of:
-
1 or 2 nucleotides shifts the codon grouping
-
-
All downstream codons are altered
Causes of Frame Shift Mutation
-
Intercalating agents (e.g., acridine dyes, ethidium bromide)
-
Errors during DNA replication
-
DNA damage and faulty repair
-
Transposable elements (jumping genes)
Molecular Mechanism
-
Insertion/deletion of nucleotide(s)
-
Shift in reading frame
-
Formation of new codons
-
Introduction of premature stop codon
-
Production of truncated or nonfunctional protein
Types of Frame Shift Mutations
1. Insertion Frame Shift
-
Addition of extra nucleotide(s)
-
Alters downstream amino acid sequence
2. Deletion Frame Shift
-
Loss of nucleotide(s)
-
Disrupts normal protein synthesis
Effects of Frame Shift Mutation
-
Completely changes amino acid sequence after mutation site
-
Produces:
-
Nonfunctional proteins
-
Truncated proteins
-
-
Usually more severe than point mutations
Example
Original sequence:
AUG-AAA-GGC-UCU
(Met-Lys-Gly-Ser)
Insertion of one base:
AUG-AUAA-AGG-CUC-U
→ altered codons → premature stop
Clinical Significance
-
Common cause of genetic disorders
-
Example:
-
Duchenne muscular dystrophy
-
Certain forms of cystic fibrosis
-
-
Often lethal or severely debilitating
Comparison: Frame Shift vs Point Mutation
| Feature | Frame Shift Mutation | Point Mutation |
|---|---|---|
| Cause | Insertion/deletion | Base substitution |
| Reading frame | Shifted | Unchanged |
| Severity | High | Variable |
| Protein effect | Severe | Mild to severe |
Physical Mutagens
Physical mutagens are physical agents, mainly radiations, that induce mutations by causing structural damage to DNA. These mutagens alter DNA either directly or indirectly through the formation of reactive molecules.
Types of Physical Mutagens
1. Ultraviolet (UV) Radiation
Source
-
Sunlight
-
UV lamps
Wavelength
-
Most mutagenic: UV-B (280–320 nm)
Mechanism of Action
-
Causes formation of pyrimidine dimers, especially:
-
Thymine–thymine dimers
-
-
Distorts DNA helix
-
Blocks:
-
DNA replication
-
Transcription
-
Consequences
-
Base substitution mutations
-
Replication errors
-
Cell death if unrepaired
DNA Repair
-
Photoreactivation
-
Nucleotide excision repair
Clinical Importance
-
Major cause of:
-
Skin cancer
-
Photoaging
-
2. Ionizing Radiation
Includes:
-
X-rays
-
Gamma rays
-
Cosmic rays
-
Alpha and beta particles
Mechanism of Action
Ionizing radiation damages DNA by:
a. Direct Effect
-
Breaks DNA strands directly
-
Causes:
-
Single-strand breaks
-
Double-strand breaks
-
b. Indirect Effect
-
Ionizes water molecules
-
Produces free radicals (•OH, •H)
-
Free radicals damage DNA bases and backbone
Types of DNA Damage
-
Base modifications
-
DNA strand breaks
-
Chromosomal deletions
-
Translocations and inversions
Biological Effects
-
Point mutations
-
Chromosomal aberrations
-
Cell death
-
Cancer development
3. Heat (Less Common)
-
High temperature can increase:
-
Depurination
-
Deamination
-
-
Acts as a weak physical mutagen
Comparison: UV vs Ionizing Radiation
| Feature | UV Radiation | Ionizing Radiation |
|---|---|---|
| Energy | Low | High |
| Main damage | Thymine dimers | Strand breaks |
| Penetration | Low | High |
| DNA breaks | Rare | Common |
| Carcinogenic | Yes | Yes |
Sources of Exposure
-
Sunlight
-
Medical imaging (X-rays, CT scans)
-
Radiotherapy
-
Nuclear accidents
-
Cosmic radiation
Cellular Defense Against Physical Mutagens
-
DNA repair mechanisms
-
Antioxidant enzymes
-
Cell cycle checkpoints
-
Apoptosis of severely damaged cells
Medical and Biological Significance
1. Medical Importance
-
Cause of:
-
Cancer
-
Genetic disorders
-
Birth defects
-
2. Research Importance
-
Used to:
-
Induce mutations experimentally
-
Study DNA repair pathways
-
3. Environmental Importance
-
Radiation exposure is a public health concern
-
Occupational hazards in healthcare and nuclear industries
Chemical Mutagens
Chemical mutagens are substances that cause heritable changes in DNA by modifying nucleotide bases, altering base-pairing properties, or disturbing DNA structure. They are an important cause of induced mutations and are widely used in genetic research as well as implicated in carcinogenesis.
Classification of Chemical Mutagens
1. Base Analogs
These chemicals resemble normal DNA bases and can be incorporated into DNA during replication.
Examples
-
5-Bromouracil (5-BU) – analog of thymine
-
2-Aminopurine – analog of adenine
Mechanism
-
Base analogs undergo tautomeric shifts
-
Pair incorrectly during replication
-
Cause transition mutations
2. Deaminating Agents
These agents remove amino groups from nitrogenous bases.
Example
-
Nitrous acid (HNO₂)
Deamination Reactions
-
Cytosine → Uracil
-
Adenine → Hypoxanthine
-
Guanine → Xanthine
Result
-
Leads mainly to transition mutations
3. Alkylating Agents
These agents add alkyl groups (–CH₃, –C₂H₅) to DNA bases.
Examples
-
Ethyl methanesulfonate (EMS)
-
Methyl methanesulfonate (MMS)
-
Mustard gas
Mechanism
-
Alkylation alters base-pairing
-
Causes:
-
Mis-pairing
-
Cross-linking
-
DNA strand breaks
-
4. Intercalating Agents
These are planar molecules that insert between base pairs of DNA.
Examples
-
Acridine dyes
-
Proflavin
-
Ethidium bromide
Mechanism
-
Distort DNA structure
-
Cause:
-
Insertions
-
Deletions
-
Result
-
Frame-shift mutations
5. Hydroxylating Agents
-
Modify bases by adding hydroxyl groups
-
Cause abnormal base pairing
Table: Chemical Mutagens
| Type | Example | Main Mutation |
|---|---|---|
| Base analogs | 5-Bromouracil | Transition |
| Deaminating agents | Nitrous acid | Transition |
| Alkylating agents | EMS | Base substitution |
| Intercalating agents | Acridine dyes | Frameshift |
Molecular Effects of Chemical Mutagens
-
Alter hydrogen bonding
-
Disturb DNA replication
-
Inhibit DNA repair
-
Increase mutation frequency
Biological and Medical Significance
1. Medical Importance
-
Cause of:
-
Genetic disorders
-
Cancer
-
Birth defects
-
2. Research Applications
-
Induction of mutations for:
-
Gene mapping
-
Functional studies
-
-
Used in Ames test to assess mutagenicity
3. Industrial Applications
-
Strain improvement
-
Enhanced production of antibiotics and enzymes
Molecular Basis of Mutation
The molecular basis of mutation refers to the changes at the DNA level that alter the genetic information of an organism. These changes may involve single nucleotide substitutions, insertions or deletions, or structural alterations of DNA, ultimately affecting gene expression and protein function.
1. Base Substitution
Base substitution involves replacement of one nucleotide base by another.
A. Transition
-
Substitution between bases of the same class
-
Purine ↔ Purine (A ↔ G)
-
Pyrimidine ↔ Pyrimidine (C ↔ T)
-
B. Transversion
-
Substitution between bases of different classes
-
Purine ↔ Pyrimidine (A or G ↔ C or T)
-
Effects of Base Substitution:
-
Silent mutation – no change in amino acid
-
Missense mutation – altered amino acid
-
Nonsense mutation – premature stop codon
2. Tautomeric Shifts of Bases
-
Nitrogenous bases exist in keto–enol or amino–imino forms
-
Rare tautomeric forms pair abnormally during replication
-
Leads to incorrect base pairing and point mutations
3. Depurination
-
Loss of purine bases (adenine or guanine) from DNA
-
Creates an apurinic (AP) site
-
DNA polymerase may:
-
Insert wrong base
-
Skip the site → mutation
-
4. Deamination
Removal of amino groups from bases:
-
Cytosine → Uracil
-
Adenine → Hypoxanthine
-
Guanine → Xanthine
Leads to transition mutations if unrepaired.
5. Insertions and Deletions (Indels)
-
Addition or loss of nucleotides
-
If not in multiples of three → frameshift mutation
-
Alters:
-
Reading frame
-
Downstream amino acid sequence
-
6. Replication Errors
-
DNA polymerase may incorporate incorrect nucleotides
-
Proofreading (3′→5′ exonuclease activity) usually corrects errors
-
Failure results in permanent mutation
7. DNA Damage by Mutagens
Physical Mutagens
-
UV radiation → thymine dimers
-
Ionizing radiation → strand breaks
Chemical Mutagens
-
Base analogs
-
Alkylating agents
-
Deaminating agents
-
Intercalating agents
8. DNA Repair Failure
Mutations persist when DNA repair mechanisms fail.
Important Repair Pathways
-
Mismatch repair
-
Base excision repair
-
Nucleotide excision repair
-
Double-strand break repair
Defects in repair pathways greatly increase mutation rates.
9. Role of Transposable Elements
-
Mobile DNA segments that move within genome
-
Cause:
-
Gene disruption
-
Insertions and deletions
-
-
Important source of spontaneous mutation
10. Molecular Consequences of Mutation
-
Altered protein structure
-
Loss or gain of function
-
Truncated proteins
-
Abnormal regulation of gene expression
Clinical Significance
-
Cause of genetic disorders
-
Oncogene activation and tumor suppressor gene inactivation
-
Development of antibiotic resistance
Site-Directed Mutagenesis
Site-directed mutagenesis is a molecular biology technique used to introduce specific, predetermined nucleotide changes at a defined site in a gene. Unlike random mutagenesis, SDM allows precise alteration of DNA to study gene and protein function.
Principle of Site-Directed Mutagenesis
-
A synthetic oligonucleotide primer containing the desired mutation is designed.
-
The primer is complementary to the target DNA except for the mutated base(s).
-
During DNA synthesis, the primer incorporates the mutation into the newly synthesized strand.
-
The mutated DNA is amplified and expressed.
Types of Site-Directed Mutagenesis
1. Substitution Mutagenesis
-
One base pair is replaced by another
-
Produces:
-
Silent mutation
-
Missense mutation
-
Nonsense mutation
-
2. Insertion Mutagenesis
-
One or more nucleotides are inserted
-
May cause:
-
Addition of amino acids
-
Frame shift mutation
-
3. Deletion Mutagenesis
-
One or more nucleotides are removed
-
Used to study functional domains of proteins
Methods of Site-Directed Mutagenesis
1. Oligonucleotide-Directed Mutagenesis (Classical Method)
-
Single-stranded DNA template used
-
Mutagenic primer introduced
-
DNA polymerase synthesizes mutated strand
-
Parental strand removed enzymatically
2. PCR-Based Site-Directed Mutagenesis (Most Common)
-
Uses PCR amplification
-
Primers contain desired mutation
-
Amplifies entire plasmid
-
Parental (methylated) DNA removed using DpnI enzyme
3. Overlap Extension PCR
-
Two PCR reactions create overlapping fragments
-
Mutation introduced in overlapping region
-
Fragments fused in final PCR
Enzymes Used
-
DNA polymerase (high-fidelity)
-
DpnI – digests methylated parental DNA
-
DNA ligase (if required)
Applications of Site-Directed Mutagenesis
Research Applications
-
Identification of active sites in enzymes
-
Studying protein-protein interactions
-
Structure-function analysis of proteins
Medical Applications
-
Understanding disease-causing mutations
-
Studying oncogenes and tumor suppressor genes
-
Drug target validation
Industrial & Biotechnological Applications
-
Enzyme engineering for:
-
Higher stability
-
Increased activity
-
Altered substrate specificity
-
-
Development of improved industrial strains
Advantages
-
Highly specific and precise
-
Reproducible
-
Minimal unwanted mutations
Isolation of Mutants
Steps in Isolation of Mutants
-
Induction of mutation (spontaneous or induced using mutagens)
-
Expression period – time allowed for mutant phenotype to appear
-
Selection or screening of mutants
-
Confirmation and characterization of mutants
Methods of Isolation of Mutants
1. Selection Methods
Selection allows only mutants to survive, while wild-type cells are eliminated.
a. Positive Selection
-
Mutants grow in presence of a selective agent
-
Wild-type cells are killed or inhibited
Examples:
-
Antibiotic-resistant mutants
-
Heavy-metal-resistant mutants
b. Negative Selection
-
Mutants fail to grow under selective conditions
-
Wild-type cells survive
Example:
-
Auxotrophic mutants
2. Screening Methods
Screening requires individual examination of colonies to identify mutants.
a. Replica Plating (Lederberg Technique)
-
Colonies from a master plate are transferred to multiple plates
-
Growth patterns compared
-
Identifies:
-
Nutritional mutants
-
Conditional mutants
-
Advantage:
Non-destructive method
b. Indicator Media Screening
-
Media containing indicators change color
-
Used to detect:
-
Sugar fermentation mutants
-
Enzyme-deficient mutants
-
3. Enrichment Techniques
Penicillin Enrichment Method
-
Penicillin kills only actively growing cells
-
Auxotrophic mutants survive due to lack of growth
-
Increases proportion of mutants
Types of Mutants Isolated
1. Auxotrophic Mutants
-
Require additional nutrients
-
Useful in metabolic pathway studies
2. Conditional Mutants
-
Express mutant phenotype only under restrictive conditions
-
Temperature-sensitive mutants are common
3. Resistant Mutants
-
Resistant to antibiotics, drugs, or toxins
-
Important in clinical microbiology
Confirmation of Mutants
-
Re-testing on selective and non-selective media
-
Genetic stability testing
-
Molecular confirmation (PCR, sequencing)
Significance of Isolation of Mutants
Research Significance
-
Identification of gene function
-
Mapping of biochemical pathways
Industrial Significance
-
Strain improvement
-
Increased yield of:
-
Antibiotics
-
Enzymes
-
Vitamins
-
Medical Significance
-
Understanding drug resistance
-
Studying pathogenic mechanisms
Ames Test
The Ames test is a biological assay used to detect the mutagenic potential of chemical substances using specially mutated strains of Salmonella typhimurium. Since most mutagens are also carcinogens, the test is widely used to assess cancer-causing potential of chemicals.
Principle of Ames Test
-
Uses histidine-dependent (his⁻) mutant strains of Salmonella typhimurium
-
These bacteria cannot grow on histidine-free media
-
When exposed to a mutagen, reverse mutation (his⁻ → his⁺) may occur
-
Revertant bacteria regain ability to synthesize histidine
-
Number of revertant colonies = degree of mutagenicity
More revertant colonies → stronger mutagen
Test Organism
-
Salmonella typhimurium
-
Genetically modified strains:
-
Unable to synthesize histidine
-
Increased permeability of cell wall
-
Defective DNA repair mechanisms (↑ sensitivity)
-
Types of Mutations Detected
Different strains are used to detect different mutations:
| Strain | Detects |
|---|---|
| TA98 | Frameshift mutations |
| TA100 | Base-pair substitutions |
| TA1535 | Point mutations |
| TA1537 | Frameshift mutations |
Role of S9 Mix (Metabolic Activation System)
-
Many chemicals are not mutagenic by themselves
-
They become mutagenic only after metabolism in the liver
-
S9 mix:
-
Prepared from rat liver microsomal enzymes
-
Simulates mammalian metabolism
-
-
Helps detect pro-mutagens
Procedure
-
Prepare histidine-dependent Salmonella strains
-
Mix bacteria with:
-
Test chemical
-
With or without S9 mix
-
-
Plate mixture on histidine-deficient agar
-
Incubate at 37°C for 24–48 hours
-
Count revertant colonies
Interpretation of Results
-
Negative Test:
-
Few revertant colonies (similar to control)
-
-
Positive Test:
-
Significant increase in revertant colonies compared to control
-
-
Dose-dependent increase strongly suggests mutagenicity
Controls Used
-
Negative control: No mutagen added
-
Positive control: Known mutagen added
-
Ensures test validity
Significance of Ames Test
1. Medical Importance
-
Screening of potential carcinogens
-
Understanding mutation-related diseases
2. Pharmaceutical Industry
-
Drug safety testing
-
Toxicity screening before human trials
3. Environmental Safety
-
Testing industrial chemicals
-
Monitoring pollutants and pesticides
4. Research Importance
-
Studying mechanisms of mutagenesis
-
Comparing mutagenic strength of compounds
Advantages
-
Simple and rapid
-
Cost-effective
-
Highly sensitive
-
Strong correlation with carcinogenicity
Limitations
-
Not all carcinogens are mutagens
-
False positives possible
-
Bacterial system ≠ human system
-
Requires metabolic activation system
MCQs
1. Mutation is best defined as:
A. Temporary change in phenotype
B. Heritable change in DNA sequence
C. Change in protein folding only
D. Change in RNA transcription rate
✅ Answer: B
2. Mutations that arise without any external agent are called:
A. Induced mutations
B. Reverse mutations
C. Spontaneous mutations
D. Conditional mutations
✅ Answer: C
3. Most spontaneous mutations occur due to:
A. Radiation
B. Replication errors
C. Antibiotics
D. Viruses
✅ Answer: B
4. Tautomeric shift causes mutation by:
A. DNA strand break
B. Chromosomal loss
C. Abnormal base pairing
D. Protein denaturation
✅ Answer: C
5. Depurination results in loss of:
A. Pyrimidine base
B. Ribose sugar
C. Purine base
D. Phosphate group
✅ Answer: C
6. Mutations caused by external agents are called:
A. Spontaneous
B. Natural
C. Induced
D. Conditional
✅ Answer: C
7. Which mutation does NOT change amino acid sequence?
A. Missense
B. Nonsense
C. Silent
D. Frameshift
✅ Answer: C
8. Silent mutation occurs due to:
A. Frame shift
B. Genetic code degeneracy
C. Protein truncation
D. RNA splicing error
✅ Answer: B
9. A frameshift mutation is caused by:
A. Base substitution
B. Deletion of three bases
C. Insertion of two bases
D. Transition mutation
✅ Answer: C
10. Frameshift mutations are usually:
A. Harmless
B. Mild
C. Severe
D. Reversible
✅ Answer: C
11. Transition mutation involves:
A. Purine → pyrimidine
B. Pyrimidine → purine
C. Purine → purine
D. Deletion of bases
✅ Answer: C
12. Transversion mutation involves:
A. Purine ↔ pyrimidine
B. Purine ↔ purine
C. Pyrimidine ↔ pyrimidine
D. Codon deletion
✅ Answer: A
13. UV radiation primarily causes:
A. DNA alkylation
B. Thymine dimers
C. Base deamination
D. DNA methylation
✅ Answer: B
14. Ionizing radiation causes:
A. Thymine dimers
B. Single-strand breaks only
C. Double-strand DNA breaks
D. Base analog incorporation
✅ Answer: C
15. Which is a physical mutagen?
A. Nitrous acid
B. EMS
C. UV radiation
D. Acridine dye
✅ Answer: C
16. 5-Bromouracil is an example of:
A. Alkylating agent
B. Intercalating agent
C. Base analog
D. Deaminating agent
✅ Answer: C
17. Nitrous acid causes mutation by:
A. Intercalation
B. Alkylation
C. Deamination
D. Cross-linking
✅ Answer: C
18. Acridine dyes cause:
A. Transitions
B. Transversions
C. Frameshift mutations
D. Point mutations
✅ Answer: C
19. EMS (ethyl methanesulfonate) is:
A. Physical mutagen
B. Alkylating agent
C. Base analog
D. Enzyme inhibitor
✅ Answer: B
20. Molecular basis of mutation includes:
A. Only base substitution
B. Only frameshift
C. Base substitution, insertion & deletion
D. Protein folding errors
✅ Answer: C
21. DNA repair failure leads to:
A. Reduced mutation rate
B. Increased mutation accumulation
C. Protein degradation
D. RNA instability
✅ Answer: B
22. Mismatch repair corrects:
A. Thymine dimers
B. Replication errors
C. Double-strand breaks
D. Alkylation damage
✅ Answer: B
23. Site-directed mutagenesis is used to:
A. Delete whole genes
B. Introduce random mutations
C. Introduce specific mutations
D. Detect carcinogens
✅ Answer: C
24. Site-directed mutagenesis uses:
A. Restriction enzymes only
B. Synthetic oligonucleotides
C. UV radiation
D. Mutagenic chemicals
✅ Answer: B
25. Which is NOT an application of site-directed mutagenesis?
A. Protein function analysis
B. Enzyme engineering
C. Antibiotic sensitivity testing
D. Gene structure study
✅ Answer: C
26. Mutagenesis is biologically significant because it:
A. Causes disease only
B. Stops evolution
C. Creates genetic variation
D. Prevents adaptation
✅ Answer: C
27. Mutations are essential for:
A. Cell death
B. Evolution
C. DNA repair
D. Protein synthesis
✅ Answer: B
28. Antibiotic resistance in bacteria arises due to:
A. Gene deletion
B. Mutagenesis
C. Protein denaturation
D. RNA degradation
✅ Answer: B
29. Industrial mutagenesis is used for:
A. Disease diagnosis
B. Vaccine development
C. Strain improvement
D. Cancer treatment
✅ Answer: C
30. Replica plating is used for:
A. DNA sequencing
B. Isolation of mutants
C. Protein purification
D. PCR amplification
✅ Answer: B
31. Screening involves:
A. Killing unwanted cells
B. Identifying mutants by phenotype
C. Selecting only wild type
D. Removing plasmids
✅ Answer: B
32. Selection technique allows:
A. Growth of all cells
B. Growth of only mutants
C. Identification by color
D. Protein isolation
✅ Answer: B
33. Ames test detects:
A. Antibiotic resistance
B. Mutagenic potential
C. Viral infection
D. DNA repair
✅ Answer: B
34. Ames test uses:
A. E. coli
B. Salmonella typhimurium
C. Bacillus subtilis
D. Yeast
✅ Answer: B
35. Ames test strains are deficient in:
A. Tryptophan synthesis
B. Histidine synthesis
C. Methionine synthesis
D. Leucine synthesis
✅ Answer: B
36. Revertant colonies indicate:
A. Cell death
B. DNA damage
C. Reverse mutation
D. Protein denaturation
✅ Answer: C
37. S9 mix in Ames test represents:
A. Bacterial enzyme
B. Plant extract
C. Liver microsomal enzymes
D. Antibiotic mixture
✅ Answer: C
38. Purpose of S9 mix is to:
A. Kill bacteria
B. Increase mutation rate
C. Simulate mammalian metabolism
D. Repair DNA
✅ Answer: C
39. Many carcinogens are also:
A. Antibiotics
B. Mutagens
C. Hormones
D. Vitamins
✅ Answer: B
40. Ames test is mainly used in:
A. Clinical diagnosis
B. Drug safety testing
C. Vaccine production
D. Gene therapy
✅ Answer: B
41. Which mutation produces a stop codon?
A. Silent
B. Missense
C. Nonsense
D. Frameshift
✅ Answer: C
42. Nonsense mutation results in:
A. Longer protein
B. Shortened protein
C. Same protein
D. No protein synthesis
✅ Answer: B
43. Transposable elements cause mutation by:
A. Base substitution
B. Moving within genome
C. DNA repair
D. Protein modification
✅ Answer: B
44. Mutations occurring in germ cells are:
A. Non-heritable
B. Somatic
C. Heritable
D. Temporary
✅ Answer: C
45. Somatic mutations affect:
A. Offspring
B. Germ line
C. Individual only
D. Evolution
✅ Answer: C
46. Which mutation is most dangerous?
A. Silent
B. Missense
C. Frameshift
D. Neutral
✅ Answer: C
47. Chemical mutagens mainly act by:
A. Breaking chromosomes
B. Modifying DNA bases
C. Inhibiting enzymes
D. Blocking ribosomes
✅ Answer: B
48. Mutation rate can be increased by:
A. DNA repair
B. Antioxidants
C. Mutagens
D. Proofreading
✅ Answer: C
49. Proofreading activity occurs in:
A. RNA polymerase
B. DNA ligase
C. DNA polymerase
D. Restriction enzyme
✅ Answer: C
50. Final outcome of mutation may be:
A. Always harmful
B. Always beneficial
C. Neutral, harmful, or beneficial
D. Always lethal
✅ Answer: C