- Antimicrobial susceptibility testing for Mycobacterium tuberculosis (M. tuberculosis) and other mycobacteria plays a critical role in identifying drug resistance patterns and guiding the treatment of tuberculosis (TB) and other mycobacterial infections.
- Given that mycobacteria, particularly M. tuberculosis, grow slowly and have complex cell wall structures, testing methods must be reliable and effective in detecting drug resistance on time.
- The emergence of multi-drug-resistant tuberculosis (MDR-TB) and extensively drug-resistant tuberculosis (XDR-TB) has made AST even more crucial in modern clinical practice.
- AST methods for Mycobacterium can be divided into phenotypic methods (which measure bacterial growth in the presence of drugs) and genotypic methods (which identify mutations in resistance-related genes).
Phenotypic Methods
Phenotypic methods involve detecting changes in bacterial growth in the presence of antimicrobial agents, indicating the bacteria’s resistance to specific drugs. These methods are based on the principle that bacteria require a certain drug concentration for growth. If they can grow despite an antibiotic, they are considered resistant.
Broth Microdilution Method (Automated Systems)
Principle:
In broth microdilution, a bacterial suspension is exposed to serially diluted concentrations of antimicrobial agents, and the minimum inhibitory concentration (MIC) is determined—the lowest concentration of the drug that prevents visible bacterial growth.
Procedure:
- A suspension of M. tuberculosis is prepared from a cultured colony and standardized to a particular density (usually 0.5 McFarland standard).
- The bacterial suspension is then added to a series of wells containing decreasing concentrations of an antibiotic.
- After incubation (typically 7-14 days for M. tuberculosis due to its slow growth), the MIC is determined by inspecting the wells for growth. The lowest concentration where no growth occurs is the MIC.
Advantages:
- This method provides quantitative information (MIC) about bacterial resistance.
- It can be automated, speeding up the process.
- It can be used to test multiple drugs simultaneously.
Disadvantages:
- It requires a pure culture of bacteria, which can take time to obtain.
- For M. tuberculosis, results can take several weeks due to slow growth.
Drugs Tested:
- First-line: Rifampicin (RIF), Isoniazid (INH), Pyrazinamide (PZA), Ethambutol (EMB)
- Second-line: Fluoroquinolones (e.g., Levofloxacin, Moxifloxacin), Aminoglycosides (e.g., Amikacin, Kanamycin), Linezolid, Capreomycin
Agar Proportion Method
Principle:
The agar proportion method is a solid-phase test where the ability of M. tuberculosis to grow in the presence of antibiotics is assessed. The bacterial inoculum is streaked on agar plates containing different concentrations of antibiotics, and the proportion of growth is used to determine resistance.
Procedure:
- A standardized suspension of M. tuberculosis is inoculated onto Lowenstein-Jensen (LJ) or Middlebrook 7H10/7H11 agar plates.
- Plates are supplemented with antibiotics at different concentrations.
- The plates are incubated for up to 4 weeks at 37°C, and the growth of colonies is observed.
- The proportion of colonies growing in the presence of the drug is compared with the growth on a control plate (no antibiotic). A resistance is identified if more than 1% of the colonies grow on the drug-supplemented plate compared to the control.
Advantages:
- A reliable method for detecting resistance in M. tuberculosis.
- It can be used for testing first-line and some second-line drugs.
- It is a widely recognized standard for resistance detection.
Disadvantages:
- It is time-consuming, as the bacterial growth takes several weeks.
- It requires solid media and skilled technicians to interpret results.
Drugs Tested:
- First-line drugs: Rifampicin, Isoniazid, Ethambutol, Pyrazinamide.
- Second-line drugs: Fluoroquinolones, Aminoglycosides, and injectable agents.
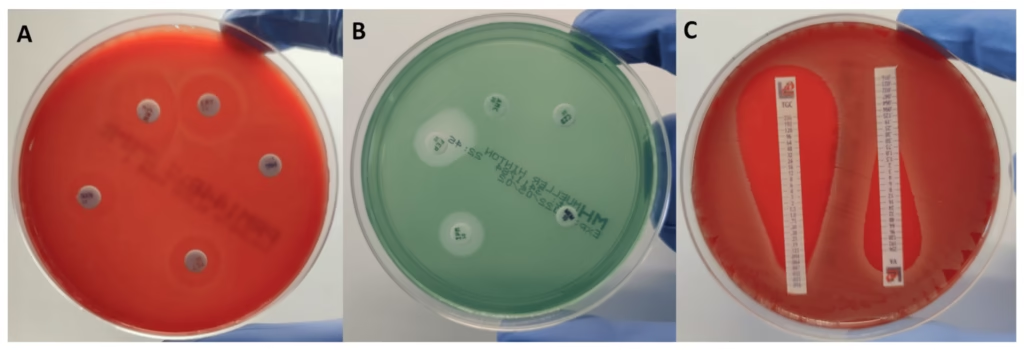
Disk Diffusion Method (Kirby-Bauer Method)
Principle:
The disk diffusion method also called the Kirby-Bauer method, is a qualitative test that assesses the susceptibility of bacteria to antibiotics based on the zone of inhibition surrounding antibiotic-impregnated disks.
Procedure:
- A lawn of M. tuberculosis is inoculated on agar plates (usually Middlebrook 7H10 or 7H11).
- Antibiotic-impregnated paper disks are placed on the inoculated agar.
- The plates are incubated, and the size of the zone of inhibition (the area with no bacterial growth) around each disk is measured.
- The size of the inhibition zone is compared to predefined standards to determine if the organism is susceptible or resistant.
Advantages:
- Simple and inexpensive.
- It can be used for screening and a limited number of drugs.
Disadvantages:
- Does not provide quantitative MIC data.
- It may not be as reliable for M. tuberculosis, which grows slowly.
- Limited by the availability of appropriate antibiotic discs.
Genotypic Methods
Genotypic methods detect mutations in the DNA of M. tuberculosis that are known to be associated with resistance. These methods are faster than phenotypic tests and do not require bacterial growth.
DNA Probes
Principle:
DNA probes are sequences of nucleotides that bind specifically to complementary sequences in the target DNA. In the case of drug resistance, DNA probes can detect mutations in genes responsible for resistance.
Procedure:
- DNA is extracted from M. tuberculosis.
- The DNA is hybridized with probes specific to resistance-associated mutations (such as those in the rpoB gene for Rifampicin resistance or the katG gene for Isoniazid resistance).
- The presence of hybridization indicates a resistant strain.
Advantages:
- Rapid detection of resistance-associated mutations.
- It can be used directly from clinical samples.
Disadvantages:
- Limited to known mutations and may miss unknown resistance mechanisms.
Applications:
- Resistance testing for Rifampicin (rpoB) and Isoniazid (katG, inhA).
Polymerase Chain Reaction (PCR)-Based Methods
Principle:
PCR amplifies specific regions of DNA, including those associated with resistance. By identifying mutations in the target genes, resistance can be determined.
Procedure:
- DNA is extracted from the mycobacterial sample.
- PCR amplification is performed on genes associated with drug resistance (such as rpoB for Rifampicin, katG for Isoniazid, or gyrA for Fluoroquinolones).
- Amplified DNA is analyzed to detect mutations associated with resistance using gel electrophoresis or fluorescent probes.
Advantages:
- Highly sensitive and specific.
- Provides results within hours to a few days.
Disadvantages:
- Only detects known mutations.
- Requires specialized equipment and expertise.
Applications:
- Rapid detection of MDR-TB (Rifampicin, Isoniazid resistance).
- Screening for XDR-TB and other drug-resistant mycobacteria.
Line Probe Assay (LPA)
Principle:
The Line Probe Assay is a PCR-based method that detects mutations in multiple genes associated with drug resistance. It uses reverse hybridization to detect amplified DNA from a patient sample.
Procedure:
- PCR amplification of target genes (e.g., rpoB, katG, inhA, gyrA).
- The PCR products are hybridized to a membrane with probes complementary to known resistance-associated mutations.
- The presence of a color change indicates the presence of specific mutations.
Advantages:
- Fast (usually 1–2 days).
- Can simultaneously test for resistance to multiple drugs (e.g., Rifampicin, Isoniazid, Fluoroquinolones).
Disadvantages:
- Requires specialized equipment and trained personnel.
- Limited to known mutations.
Applications:
- Screening for MDR-TB, XDR-TB, and drug-resistant mycobacteria.
- Used in high-burden TB settings for rapid testing.
Key Drugs for Mycobacterium Resistance Testing
The primary drugs used in the treatment of M. tuberculosis include:
- First-Line Drugs:
- Isoniazid (INH): Inhibits mycolic acid synthesis, essential for the mycobacterial cell wall.
- Rifampicin (RIF): Inhibits bacterial RNA polymerase.
- Pyrazinamide (PZA): Works under acidic conditions, targeting the M. tuberculosis cell wall.
- Ethambutol (EMB): Inhibits cell wall biosynthesis.
- Second-Line Drugs:
- Fluoroquinolones (e.g., Levofloxacin, Moxifloxacin): Inhibit DNA gyrase, affecting DNA replication.
- Aminoglycosides (e.g., Amikacin, Kanamycin): Inhibit protein synthesis by binding to bacterial ribosomes.
- Linezolid: Inhibits protein synthesis by binding to the 23S ribosomal RNA.
- Capreomycin: A second-line injectable agent with effects on protein synthesis.
Resistance Mechanisms:
- Rifampicin resistance: Mutations in the rpoB gene.
- Isoniazid resistance: Mutations in the katG, inhA, or ahpC genes.
- Fluoroquinolone resistance: Mutations in the gyrA and gyrB genes.