Introduction
- Medical microbiology is a cornerstone of clinical diagnostics, providing critical information on the detection, identification, and antimicrobial susceptibility of infectious agents.
- Historically, microbiological methods relied heavily on manual techniques: preparing culture plates, inoculating specimens, performing biochemical tests, and interpreting growth patterns by eye.

- While these methods formed the foundation of diagnostic microbiology, they suffer from limitations such as:
-
Long turnaround times (days to weeks for certain organisms).
-
Labor-intensive and repetitive manual work.
-
Variability in accuracy due to human interpretation.
-
Challenges in handling increasing specimen volumes in modern hospitals.

- Automation in medical microbiology refers to the integration of robotics, advanced imaging, molecular technologies, and artificial intelligence (AI) to replace or complement manual methods in the laboratory.
- Automation increases efficiency, reproducibility, and diagnostic accuracy, while significantly reducing turnaround times.
- It is now considered a crucial innovation to meet the growing global burden of infectious diseases and antimicrobial resistance.
Evolution of Automation in Microbiology
Pre-automation Era
-
Reliance on manual inoculation, culture, microscopy, and biochemical assays.
-
First wave of mechanization (mid-20th century): blood culture monitoring machines and semi-automated biochemical test kits.
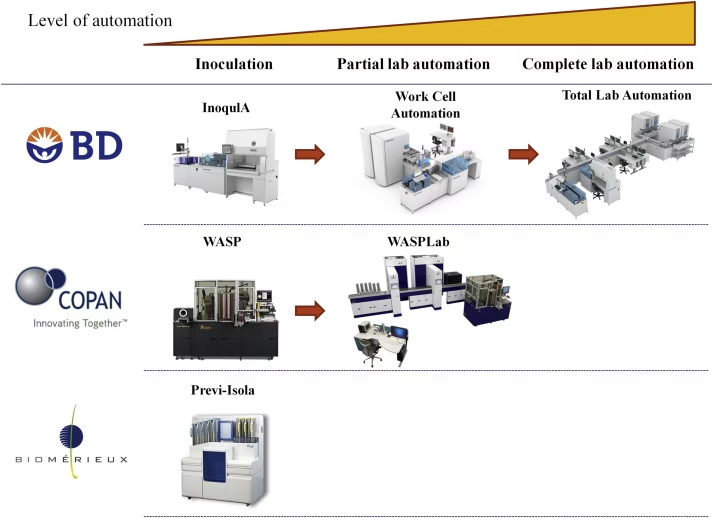
First Phase of Automation (1970s–1990s)
-
Blood culture automation (e.g., BACTEC, BacT/ALERT).
-
Automated microbial identification and susceptibility platforms (e.g., VITEK introduced in 1970s).
-
Limited scope—systems designed for specific tasks.
Second Phase (2000–2010)
-
Integration of robotics for specimen processing (plating and streaking).
-
Development of total laboratory automation (TLA) systems.
-
Emergence of MALDI-TOF MS as a rapid identification tool.
Current Era (2010 onwards)
-
Fully integrated digital laboratories with automated inoculation, incubation, imaging, and reporting.
-
Molecular automation (PCR, multiplex panels).
-
AI-driven image analysis and digital microbiology.
-
Next-generation sequencing (NGS) for genomic epidemiology and outbreak surveillance.
Major Areas of Automation in Microbiology
1. Specimen Reception and Processing
-
Automated systems manage barcoding, sorting, plating, and streaking.
-
Standardizes inoculation patterns → improved colony isolation.
-
Examples:
-
WASP (Walk-Away Specimen Processor, Copan)
-
InoqulA (BD Kiestra)
-
Colibri (Copan)
-
Impact: Reduces manual workload, ensures consistent quality, and improves biosafety by minimizing direct contact with pathogens.
2. Culture and Incubation
-
Automated incubators maintain controlled growth environments (temperature, humidity, CO₂).
-
Integrated digital cameras take high-resolution images at intervals → eliminates need for manual plate handling.
-
AI algorithms track colony growth and hemolysis patterns.
-
Systems:
-
BD Kiestra TLA
-
Copan WASPLab
-
Impact: Enables remote reading of cultures, earlier detection of growth, and digital archiving of culture images for teaching or reanalysis.
3. Microbial Identification
a. Traditional Automated Biochemical Systems
-
Use miniaturized panels for biochemical reactions.
-
Systems: VITEK 2, BD Phoenix, MicroScan WalkAway.
-
Provide organism identification in hours (vs. days by manual methods).
b. MALDI-TOF MS (Matrix-Assisted Laser Desorption Ionization – Time of Flight Mass Spectrometry)
-
Gold standard for rapid microbial identification.
-
Works by analyzing unique protein mass spectral patterns.
-
Results available in minutes, cost per test is low after setup.
-
Platforms: Bruker Biotyper, VITEK MS.
Advantages: Accurate for bacteria, yeasts, mycobacteria; useful in polymicrobial cultures with optimization.
4. Antimicrobial Susceptibility Testing (AST)
-
Determines resistance profile to guide therapy.
Automated AST platforms:
-
VITEK 2 – provides MIC values, resistance mechanisms.
-
BD Phoenix – automated broth microdilution.
-
MicroScan WalkAway – wide panels for Gram-positive and Gram-negative bacteria.
Impact: Rapid AST enables early, targeted antibiotic therapy, crucial in sepsis management.
5. Blood Culture Systems
-
Automated continuous monitoring detects microbial growth via CO₂ production or pressure changes.
-
Examples: BACTEC FX, BacT/ALERT VIRTUO, VersaTREK.
-
Reduce time to positivity → critical in sepsis.
-
Integration with MALDI-TOF allows “direct from bottle” organism ID.
6. Molecular Diagnostic Automation
-
PCR-based platforms detect pathogens and resistance genes within hours.
-
Multiplex assays test for multiple organisms simultaneously.
-
Examples:
-
FilmArray BioFire (respiratory, GI, meningitis panels)
-
Cepheid GeneXpert (TB, MRSA, COVID-19)
-
Abbott m2000, Roche cobas platforms
-
Impact: Vital in diagnosing fastidious organisms (Mycobacterium tuberculosis, viruses, C. difficile) where culture is slow or difficult.
7. Next-Generation Sequencing (NGS) and Genomics
-
Whole genome sequencing for pathogen typing and outbreak tracing.
-
Metagenomic sequencing for culture-independent pathogen detection directly from clinical samples.
-
Platforms: Illumina, Oxford Nanopore.
Applications:
-
Identifying resistance genes.
-
Tracking hospital outbreaks.
-
Studying pathogen evolution.
8. Digital Microbiology and AI
-
Automated imaging of plates combined with AI-based colony recognition.
-
Software detects colony size, morphology, color, hemolysis.
-
Can pre-screen negative plates → reduces manual review load.
-
Example: APAS Independence (LBT Innovations) uses AI to interpret culture plates.
Advantages of Automation in Microbiology
-
Improved Accuracy and Reproducibility – eliminates human variability.
-
Faster Turnaround Time – rapid pathogen identification and AST.
-
High Throughput – handles thousands of samples daily.
-
Standardization – uniform results across labs.
-
Better Infection Control – earlier detection → timely isolation.
-
Labor Efficiency – staff focus on analysis, not repetitive tasks.
-
Digital Data Archiving – permanent image storage for teaching, audits, and medico-legal purposes.
-
Enhanced Biosafety – reduced direct handling of infectious material.
Limitations and Challenges
-
High Initial Cost – installation and maintenance are expensive.
-
Database Limitations – uncommon organisms may not be identified by automated systems.
-
Infrastructure Requirements – space, power, IT support, LIS integration.
-
Training Needs – staff require advanced technical skills.
-
Over-Reliance on Automation – risk of reduced manual skills in microbiologists.
-
Accessibility – not feasible in low-resource settings.
Clinical Applications
-
Bacteriology: Rapid identification of sepsis and UTI pathogens.
-
Mycology: MALDI-TOF for Candida and Aspergillus.
-
Mycobacteriology: Automated liquid culture (MGIT 960), GeneXpert for TB.
-
Virology: PCR panels for respiratory viruses, HIV viral load monitoring.
-
Parasitology: Molecular detection of malaria, toxoplasmosis.
-
Epidemiology: NGS in outbreak tracing (e.g., foodborne Salmonella, COVID-19 variants).
Future of Automation in Microbiology
-
AI and Machine Learning: More advanced colony recognition, predictive outbreak modeling.
-
Point-of-Care Automation: Portable rapid diagnostic kits integrated with cloud reporting.
-
Microfluidics and Lab-on-a-Chip: Miniaturized, multiplexed platforms.
-
Automated Antimicrobial Stewardship: Linking AST with clinical decision support to optimize antibiotic use.
-
Integration with One Health: Automated zoonotic pathogen monitoring at the human-animal-environment interface.
-
Cloud-Based Digital Labs: Remote analysis, big data integration for global disease surveillance.
Comparison: Manual vs Automated
| Feature | Manual Methods | Automated Methods |
|---|---|---|
| Turnaround Time | Days to weeks | Hours to days |
| Labor Intensity | High | Low (robot-assisted) |
| Accuracy | Operator-dependent | High, standardized |
| Throughput | Limited | Very high |
| Cost | Low initial, high labor cost | High initial, lower labor cost long-term |
| Flexibility | Broad organism range | Limited to database/library scope |