
Introduction
Southern blotting is a method in molecular biology used to find specific DNA sequences in a sample of DNA. It was developed by Edwin M. Southern in 1975.
The method includes:
-
Cutting DNA into smaller pieces using restriction enzymes.
-
Separating these DNA fragments using agarose gel electrophoresis.
-
Moving (blotting) the DNA fragments onto a strong membrane.
-
Adding a labeled probe that sticks only to the matching DNA sequence.
-
Detecting the probe to know where and how big the DNA fragment is.
Even though modern techniques like PCR and sequencing are faster, Southern blotting is still useful to check DNA size, structure, mutations, and to confirm gene presence or cloning experiments.
Principle
Southern blotting is based on three simple ideas: 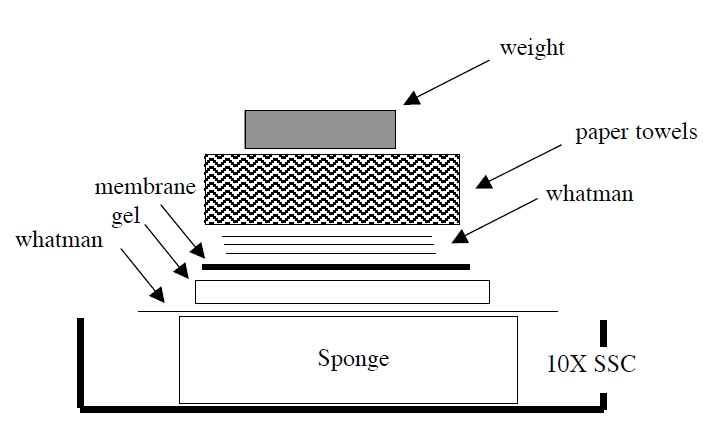
-
Cutting DNA
-
Special enzymes (restriction enzymes) cut DNA into pieces.
-
The size of the fragment depends on where the enzyme cuts.
-
-
Separating DNA by size
-
The DNA fragments are run on an agarose gel.
-
Small fragments move faster; large fragments move slower.
-
-
Finding DNA with a probe
-
DNA fragments are made single-stranded and attached to a nylon/nitrocellulose membrane.
-
A probe (a small piece of DNA with a label) is added. It sticks only to the matching DNA.
-
The label on the probe lets us see the DNA on the membrane.
-
This gives information about whether the DNA is present, how many copies exist, and the fragment size.
Materials
Equipment
-
Agarose gel system
-
UV transilluminator or light box
-
Blotting system (capillary, vacuum, or electroblotting)
-
Membranes (nylon/nitrocellulose)
-
Hybridization oven
-
Imaging system (X-ray film, chemiluminescence detector, or fluorescence scanner)
Chemicals and Reagents
-
DNA sample
-
Restriction enzymes
-
Agarose + electrophoresis buffer
-
DNA ladder (size marker)
-
Denaturation solution (NaOH + NaCl)
-
Neutralization solution (Tris-HCl + NaCl)
-
SSC buffer
-
Labeled probe (radioactive or non-radioactive)
-
Hybridization buffer
-
Washing solutions (SSC + SDS)
-
Detection chemicals or films
Steps of Southern Blotting
Step 1: DNA Extraction
-
Collect DNA from cells or tissues.
-
Check purity and amount.
Step 2: Restriction Digestion
-
Cut DNA into fragments using restriction enzymes.
-
Test a small portion on agarose gel to confirm digestion.
Step 3: Gel Electrophoresis
-
Load DNA on agarose gel and run it.
-
Stain DNA to check separation.
Step 4: Denaturation and Neutralization
-
Soak gel in alkaline solution to make DNA single-stranded.
-
Neutralize to protect DNA structure.
Step 5: Transfer (Blotting)
-
Place the gel on a platform in SSC buffer.
-
Put the membrane on top, then the paper and the weight.
-
DNA moves from the gel to the membrane overnight by capillary action.
-
Other faster methods: vacuum or electroblotting.
Step 6: Fixation
-
DNA is fixed to the membrane using UV light or baking.
Step 7: Probe Labeling
-
Make a probe that matches the target DNA.
-
Add a label (radioactive, DIG, biotin, fluorescent dye).
Step 8: Hybridization
-
Block membrane to reduce background.
-
Add probe and let it bind to matching DNA overnight.
Step 9: Washing
-
Wash with SSC/SDS buffers to remove probes that do not bind strongly.
Step 10: Detection
-
Radioactive probes → detected by X-ray film.
-
DIG/biotin probes → enzyme reactions + chemiluminescence.
-
Fluorescent probes → detected with a fluorescence scanner.
Results
-
Bands show where the probe has bound.
-
Band size tells the fragment length.
-
Band strength shows how many copies are present.
-
Multiple bands may mean there are similar genes, mutations, or different restriction sites.
Applications
-
Gene Mapping — shows where a gene is in the DNA and how many copies exist.
-
Mutation Detection — finds insertions, deletions, and changes in restriction sites.
-
Cloning and Transgenics — checks if a gene was inserted into a vector or genome.
-
RFLP Analysis — used in genetic fingerprinting.
-
Forensics — an early DNA profiling method.
-
Virus Detection — identifies viral DNA inside host cells.
-
Epigenetics — detects DNA methylation using special enzymes.
Advantages
-
Gives fragment size information.
-
Can detect gene organisation and changes.
-
Very specific if the probe is well designed.
-
Membranes can be reused with different probes.
Limitations
-
Takes 2–3 days to finish.
-
Needs large DNA amounts.
-
Not as sensitive as PCR.
-
Radioactive probes need special safety handling.
-
Labor-heavy and not good for large sample numbers.
-
Cannot detect very small mutations unless they affect restriction sites.
Troubleshooting
-
Weak signal → DNA degraded, probe not labeled well, poor transfer.
-
High background → not enough washing, poor blocking.
-
Smearing → incomplete digestion or poor DNA quality.
-
Extra bands → non-specific probe binding or rearranged DNA.
Variants
-
Dot/Slot blot → checks presence/absence only, no size info.
-
Alkaline blot → better for large DNA.
-
Methylation blot → used for epigenetic studies.