Introduction
-
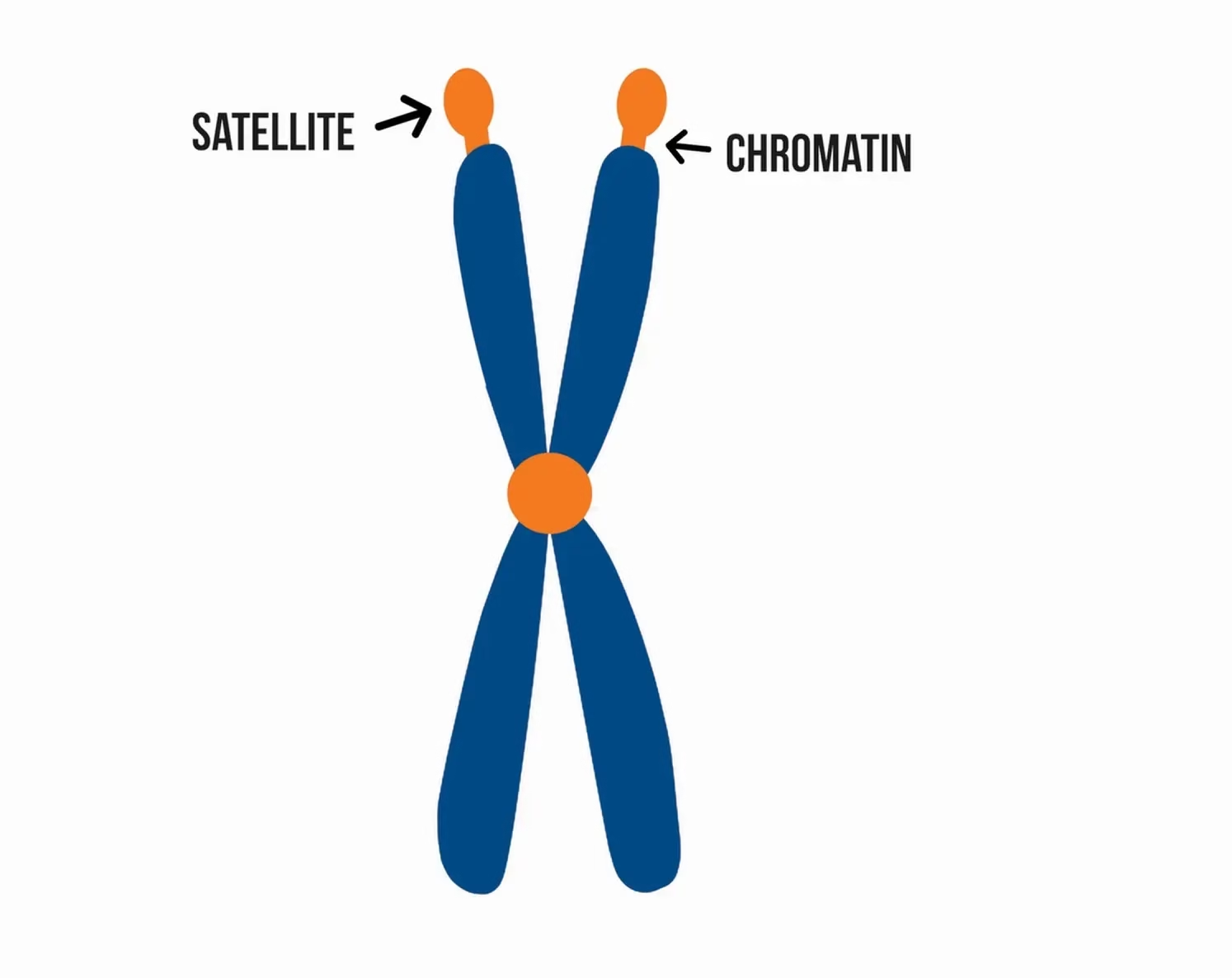
Satellite DNA refers to repetitive, non-coding DNA sequences that form distinct “satellite bands” when genomic DNA is separated by density-gradient ultracentrifugation.
-
First described by Yasmineo and colleagues (1960s) when studying mouse DNA.
-
Called “satellite” because they appear as lighter or heavier bands separate from the main DNA in cesium chloride (CsCl) gradients.
-
Present in eukaryotic genomes, often in heterochromatin, especially centromeric and pericentromeric regions.
-
Does not code for proteins but has structural, regulatory, and evolutionary roles.
General Features
-
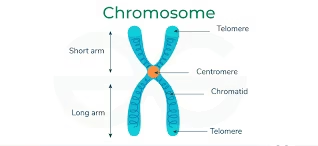
Made of tandemly repeated sequences (identical or nearly identical motifs).
-
Repeat length may range from 1 bp → several hundred bp.
-
Total length can extend up to megabases of DNA.
-
Found mostly in non-coding heterochromatic DNA.
-
Highly species-specific (useful in evolutionary studies).
-
Shows high polymorphism in length and sequence (basis of DNA profiling).
Classification
Satellite DNA is classified based on repeat unit size:
A. Classical (Major) Satellite DNA
-
Long repeats, >100 bp per unit.
-
Found in centromeres, pericentromeres.
-
Example: Human α-satellite DNA (171 bp repeat unit, organized in higher-order arrays).
B. Minisatellite DNA
-
Repeat size: 10–60 bp.
-
Length: up to 20 kb.
-
Found in subtelomeric regions, telomeres.
-
Example: Telomeric repeat (TTAGGG)n.
-
Applications: DNA fingerprinting, population genetics, identity testing.
C. Microsatellite DNA
-
Repeat size: 1–6 bp.
-
Widely distributed throughout the genome.
-
Highly polymorphic (vary between individuals).
-
Example: (CA)n, (CAG)n repeats.
-
Applications: Forensics, paternity testing, disease diagnostics.
D. Satellite-like DNA (Intermediate Repeats)
-
Interspersed repeats that resemble satellites but not strictly tandem.
-
Example: Variable Number Tandem Repeats (VNTRs).
Location in Genome
-
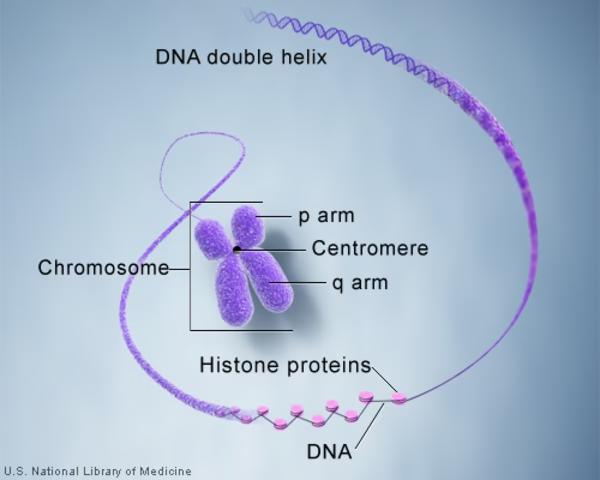
Centromeres → α-satellite DNA in humans stabilizes kinetochore assembly.
-
Telomeres → Minisatellite repeats protect chromosome ends.
-
Pericentromeric heterochromatin → structural stability.
-
Y chromosome → contains specific satellite repeats.
Molecular Organization
-
Arranged as tandem head-to-tail repeats.
-
Organized into higher-order repeats (HORs) in centromeres.
-
Shows variation in copy number (basis of genetic polymorphism).
Functions
Structural Roles
-
Centromere function: essential for kinetochore assembly & chromosome segregation.
-
Telomere stability: prevents chromosome shortening and fusion.
-
Maintains nuclear architecture (heterochromatin organization).
Regulatory Roles
-
Influences epigenetic modifications (DNA methylation, histone modification).
-
Regulates gene expression near repeat-rich regions.
Evolutionary Roles
-
Satellite DNA evolves rapidly → used as phylogenetic markers.
-
Explains genome size variability (C-value paradox).
Practical Roles
-
Used in DNA fingerprinting, forensic science, population genetics, paternity testing.
-
Helps in marker-assisted selection in plants and animals.
Techniques
-
CsCl density gradient centrifugation → classical method (separate satellite bands).
-
Restriction enzyme digestion & gel electrophoresis → reveals repeat motifs.
-
Southern blotting → detects repeat patterns.
-
FISH (Fluorescence In Situ Hybridization) → chromosomal mapping of repeats.
-
PCR amplification → microsatellite/minisatellite analysis.
-
Next-generation sequencing (NGS) → detailed repeat characterization.
Satellite DNA and Human Disease
-
Though mostly non-coding, expansions or instability of repeats can cause disease.
-
Trinucleotide Repeat Disorders (microsatellite expansions):
-
Huntington’s disease → (CAG)n repeat expansion.
-
Fragile X syndrome → (CGG)n expansion in FMR1 gene.
-
Myotonic dystrophy → (CTG)n expansion.
-
-
Genomic instability in cancer → microsatellite instability (MSI).
-
Epigenetic misregulation of satellite DNA linked to autoimmune diseases.
Applications
-
DNA fingerprinting (minisatellites, microsatellites).
-
Forensic analysis (STR profiling in crime cases).
-
Paternity testing (high polymorphism).
-
Genetic linkage studies (microsatellite markers).
-
Evolutionary biology (species-specific satellites as phylogenetic tools).
-
Medical diagnostics (detecting repeat expansion disorders).