Introduction
- Chromosomes are thread-like structures present inside the nucleus of every living cell.
- They are made up of DNA (deoxyribonucleic acid) and proteins, mainly histones, which help in packaging the long DNA molecules into a compact form.
- Each chromosome carries genes, the units of heredity, that control the traits and functions of an organism.
- In humans, chromosomes occur in pairs.
- A normal human cell has 46 chromosomes (23 pairs), out of which 22 pairs are autosomes and 1 pair are sex chromosomes (XX in females and XY in males).
- During cell division, chromosomes ensure the accurate distribution of genetic material from parent cells to daughter cells.
- Thus, chromosomes play a central role in storing, protecting, and transmitting genetic information from one generation to the next.
Structure of Chromosomes
Chromosomes are highly condensed thread-like structures composed of DNA and proteins, found in the nucleus of eukaryotic cells, carrying genetic information.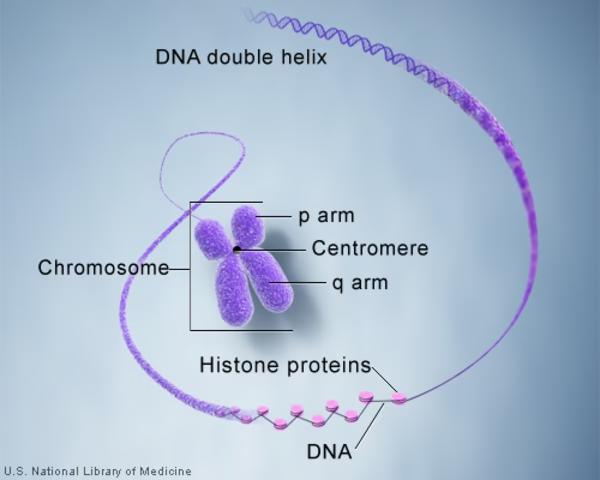
-
Chemical composition:
-
DNA (~40%): double-helical molecule containing genes.
-
Histones (~50%): basic proteins (H1, H2A, H2B, H3, H4) that package DNA into nucleosomes.
-
Non-histone proteins (~10%): enzymes (polymerases, topoisomerases), regulatory proteins, scaffold proteins.
-
RNA molecules (small amount, regulatory).
-
-
Levels of organization:
-
DNA double helix → 2 nm.
-
Nucleosomes (DNA wrapped around histone octamer) → “beads-on-a-string” form (10 nm fiber).
-
30 nm solenoid fiber → coiling of nucleosomes.
-
Chromatin loops (300 nm).
-
Condensed metaphase chromosome (1400 nm).
-
-
Morphology of a metaphase chromosome:
-
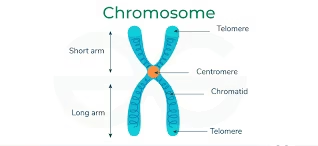
Chromatid: Each duplicated chromosome has two identical chromatids.
-
Centromere: Constriction region dividing chromosome into short arm (p) and long arm (q).
-
Telomeres: Repetitive DNA at chromosome ends (TTAGGG in humans), protecting against degradation and fusion.
-
Secondary constrictions/NOR (Nucleolar Organizer Regions): Sites of ribosomal RNA (rRNA) synthesis.
-
Satellite bodies: Small chromatin masses attached to secondary constrictions.
-
-
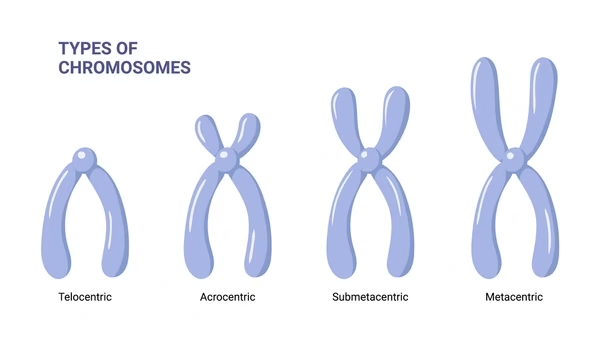
Types of chromosomes (based on centromere position):
-
Metacentric: centromere in middle.
-
Submetacentric: centromere slightly off-center.
-
Acrocentric: centromere near one end (humans: 13,14,15,21,22).
-
Telocentric: centromere at extreme end (not in humans, common in rodents).
-
Number of Chromosomes
Each species has a constant diploid (2n) chromosome number.
Examples:
-
Humans → 46 (23 pairs).
-
Chimpanzee → 48.
-
Drosophila → 8.
-
Onion → 16.
-
Dog → 78.
Diploid (2n): complete set (somatic cells).
-
Haploid (n): half set (gametes).
-
Chromosomal abnormalities:
-
Aneuploidy (loss/gain of a chromosome) – e.g., Down syndrome (Trisomy 21), Turner syndrome (45,X), Klinefelter syndrome (47,XXY).
-
Polyploidy (extra complete sets) – common in plants, rare in humans.
-
Sex Chromosomes
-
Autosomes: non-sex chromosomes (22 pairs in humans).
-
Sex chromosomes: determine biological sex (XX in female, XY in male).
-
Y chromosome: smallest human chromosome; carries SRY gene → triggers male development.
-
Sex determination:
-
XX = female.
-
XY = male.
-
-
Disorders of sex chromosomes:
-
Turner syndrome (45,X).
-
Klinefelter syndrome (47,XXY).
-
Triple X syndrome (47,XXX).
-
XYY males (47,XYY).
-
Human Karyotype
Complete set of chromosomes of an organism, arranged in homologous pairs, decreasing in size.
-
Human karyotype:
-
Normal female: 46,XX.
-
Normal male: 46,XY.
-
-
Techniques for preparation:
-
Collect dividing cells (blood lymphocytes, bone marrow, amniotic fluid).
-
Arrest at metaphase (using colchicine).
-
Hypotonic treatment (swells cells).
-
Fixation, spreading, and staining.
-
-
Uses:
-
Identify numerical abnormalities (trisomy, monosomy).
-
Identify structural abnormalities (translocations, deletions).
-
Prenatal diagnosis (amniocentesis, chorionic villus sampling).
-
Cancer diagnosis (e.g., Philadelphia chromosome in CML).
-
Methods for Chromosome Analysis
-
Conventional cytogenetics:
-
Karyotyping.
-
Banding techniques.
-
-
Molecular cytogenetics:
-
FISH (Fluorescence In Situ Hybridization).
-
CGH (Comparative Genomic Hybridization).
-
Array-CGH (microarray based).
-
-
Flow cytometry: analysis of DNA content, ploidy, and cell cycle distribution.
-
Next-generation sequencing (NGS): genome-wide chromosomal studies.
Chromosome Banding
Developed to identify each chromosome uniquely.
-
Types of banding:
-
G-banding: Giemsa stain → alternating dark/light bands. AT-rich regions appear dark.
-
Q-banding: Quinacrine → fluorescent bands.
-
R-banding: reverse of G-banding (GC-rich areas).
-
C-banding: stains centromeric heterochromatin.
-
T-banding: highlights telomeric regions.
-
-
Applications:
-
Detecting structural abnormalities (deletions, duplications, translocations).
-
Genetic counseling and prenatal testing.
-
Fluorescence In Situ Hybridization
Principle: DNA probes labeled with fluorescent dyes hybridize to complementary chromosome regions.
-
Types of probes:
-
Locus-specific probes (for single genes).
-
Centromere-specific probes (detect aneuploidy).
-
Whole-chromosome painting probes (translocations).
-
-
Applications:
-
Detect microdeletions (e.g., DiGeorge syndrome 22q11).
-
Detect oncogene amplification (HER2 in breast cancer).
-
Identify cryptic chromosomal rearrangements.
-
Rapid prenatal diagnosis of trisomies.
-
Comparative Genomic Hybridization
Test DNA (patient) and reference DNA are labelled with different fluorescent dyes and hybridised to normal metaphase chromosomes or DNA microarrays.
-
Applications:
-
Detect genome-wide copy number variations (CNVs).
-
Detect gains/losses in tumor cells.
-
Array-CGH allows detection of very small deletions/duplications.
-
-
Limitations:
-
Cannot detect balanced rearrangements (translocations, inversions).
-
Requires specialized equipment.
-
Flow Cytometry
Cells stained with a DNA-binding fluorescent dye pass through a laser beam. Fluorescence intensity ∝ DNA content.
-
Applications:
-
Cell cycle analysis (proportion of G0/G1, S, G2/M cells).
-
Detect aneuploidy and polyploidy.
-
Immunophenotyping (with antibodies).
-
Used in oncology (tumor DNA content, prognosis).
-
Cell Cycle
-
Phases:
-
G1 phase: cell growth, protein synthesis.
-
S phase: DNA replication, centrosome duplication.
-
G2 phase: preparation for mitosis, repair of replication errors.
-
M phase: mitosis (prophase, metaphase, anaphase, telophase) + cytokinesis.
-
G0 phase: resting stage (non-dividing cells like neurons, muscle).
-
-
Regulation:
-
Controlled by cyclins and CDKs.
-
Checkpoints:
-
G1/S (DNA damage check).
-
G2/M (DNA replication completion).
-
Spindle checkpoint (chromosome alignment).
-
-
-
Dysregulation → cancer.
Mitosis
-
Purpose: Growth, repair, asexual reproduction.
-
Produces 2 identical diploid daughter cells.
-
Phases:
-
Prophase: chromosomes condense, spindle forms.
-
Metaphase: chromosomes align at equator.
-
Anaphase: sister chromatids separate.
-
Telophase: nuclear envelope reforms.
-
Cytokinesis: cytoplasm divides.
-
-
Significance:
-
Maintains genetic stability.
-
Errors may lead to cancer.
-
Meiosis
-
Purpose: Gamete formation, sexual reproduction.
-
Produces 4 haploid cells, genetically different.
-
Meiosis I (reductional division):
-
Homologous chromosomes pair (synapsis).
-
Crossing over occurs (genetic recombination at chiasmata).
-
Homologs separate → 2 haploid cells.
-
-
Meiosis II (equational division):
-
Sister chromatids separate → 4 haploid gametes.
-
-
Significance:
-
Maintains chromosome number across generations.
-
Introduces genetic diversity (crossing-over, independent assortment).
-
Errors cause nondisjunction → aneuploidy.
-