Introduction
-
DNA fingerprinting is a molecular technique used to identify individuals based on unique patterns in their DNA.
-
Except for identical twins, every person has a unique DNA profile.
-
The technique is widely used in forensic science, paternity testing, criminal investigations, and population genetics.
-
DNA fingerprinting is based on the analysis of highly variable regions of DNA, known as repetitive sequences.
-
It has revolutionized the justice system by providing highly accurate biological identification.

History of DNA Fingerprinting
DNA fingerprinting was discovered in 1984 by Sir Alec Jeffreys at the University of Leicester.
Key milestones:
-
1984 – Discovery of hypervariable regions in human DNA.
-
1985 – First application in an immigration case.
-
1986 – First criminal conviction using DNA evidence.
-
1990s – Development of PCR-based STR analysis.
-
Present – Use of automated capillary electrophoresis and national DNA databases.
Principle of DNA Fingerprinting
-
Human DNA contains repetitive sequences called:
-
VNTRs (Variable Number Tandem Repeats)
-
STRs (Short Tandem Repeats)
-
-
The number of repeat units varies among individuals.
-
These variations create unique banding patterns after analysis.
-
PCR amplification allows detection from even small biological samples.
Types of Genetic Markers Used
-
VNTRs – Longer repeat sequences (used in early methods).
-
STRs – Short repeats (2–6 base pairs), currently most commonly used.
-
SNPs (Single Nucleotide Polymorphisms) – Single base variations.
-
mtDNA analysis – Used when nuclear DNA is degraded.
-
Y-chromosome markers – Useful in paternal lineage tracing.
Steps in DNA Fingerprinting
1. Sample Collection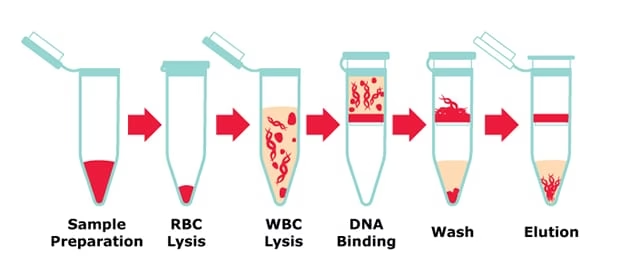
Sources of Biological Material
-
Blood stains
-
Saliva (buccal swab)
-
Semen
-
Hair root
-
Tissue
-
Bone or teeth (in decomposed bodies)
Important Considerations
-
Proper labeling
-
Use of sterile tools
-
Avoid contamination
-
Maintain chain of custody (especially in forensic cases)
2. DNA Extraction
The goal is to isolate pure DNA from cells.
Methods Used
-
Organic extraction (Phenol–chloroform method)
-
Chelex method
-
Silica column-based extraction
-
Magnetic bead technology
Steps Involved
-
Cell lysis (break open cells)
-
Protein removal
-
DNA purification
3. DNA Quantification
Before amplification, the quantity and quality of DNA must be assessed.
Methods
-
Spectrophotometry
-
Real-time PCR (most accurate)
Purpose:
-
Ensures optimal DNA concentration
-
Prevents overloading or underloading PCR
4. PCR Amplification of STR Markers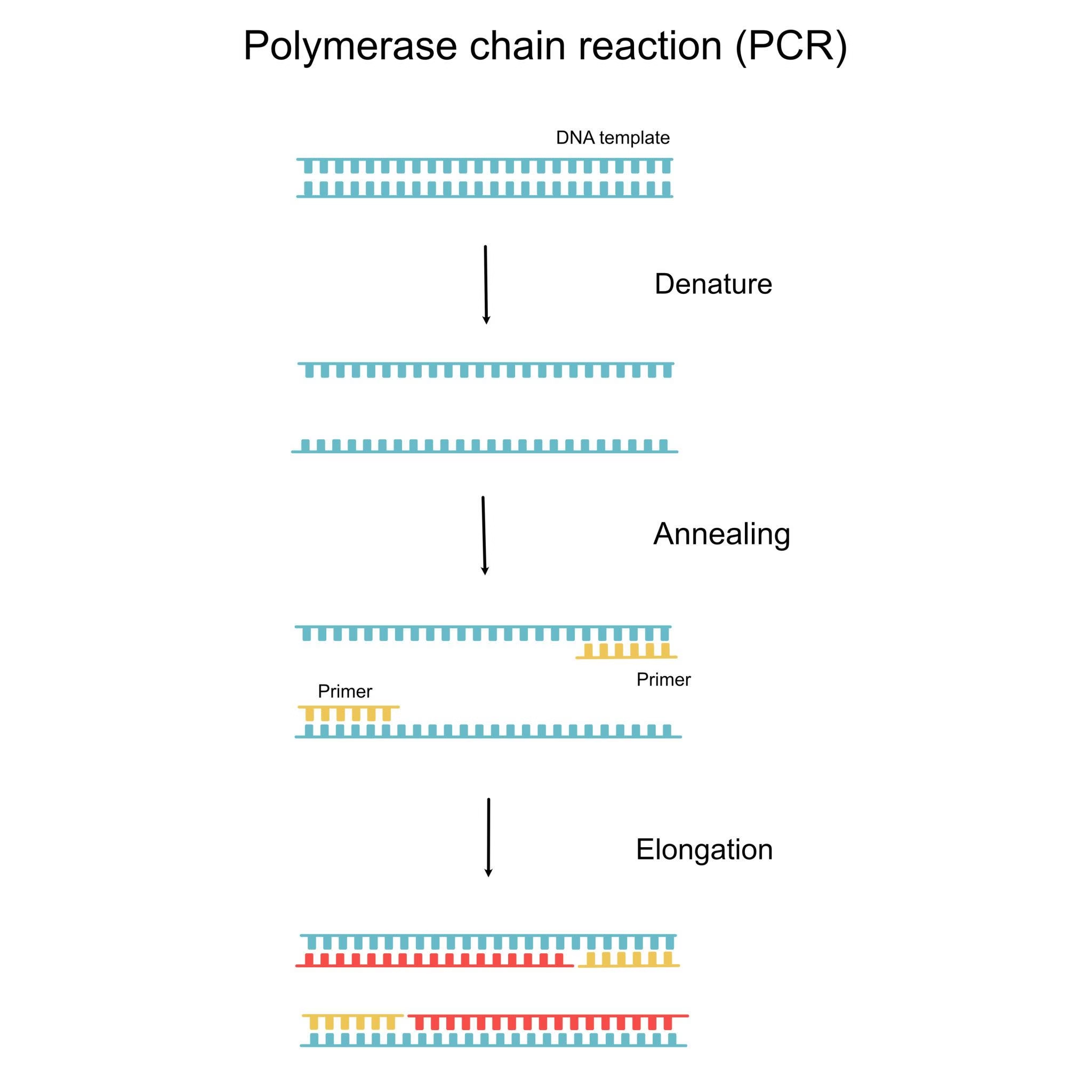
This is the most crucial step.
Principle
-
STR regions are flanked by specific primers.
-
PCR amplifies these regions exponentially.
Components of PCR
-
Template DNA
-
Primers
-
DNA polymerase (Taq polymerase)
-
dNTPs
-
Buffer solution
PCR Cycles
-
Denaturation (94–95°C)
-
Annealing (50–65°C)
-
Extension (72°C)
Multiple STR loci (usually 13–20 standard markers) are amplified simultaneously using multiplex PCR.
5. Separation of DNA Fragments
After amplification, DNA fragments are separated based on size.
Methods
A. Agarose Gel Electrophoresis (Older Method)
-
DNA fragments move through gel under electric field.
-
Smaller fragments move faster.
B. Capillary Electrophoresis (Modern Standard)
-
High precision.
-
Automated.
-
Fluorescent labeling allows digital detection.
6. Detection and Visualization
Earlier Method
-
Autoradiography showing band patterns.
Modern Method
-
Fluorescent detection.
-
Generates an electropherogram with peaks.
-
Each peak corresponds to an allele at a specific STR locus.
7. DNA Profile Generation
The final DNA profile consists of:
-
A series of numbers representing repeat counts at each STR locus.
-
Example:
Locus D3S1358 → 15, 18
Locus vWA → 16, 17
This numeric code forms an individual’s DNA profile.
8. Data Analysis and Interpretation
-
Compare DNA profile with suspect or database sample.
-
Calculate Random Match Probability (RMP).
-
Determine:
-
Match
-
Exclusion
-
Partial match
-
Statistical calculations ensure scientific reliability.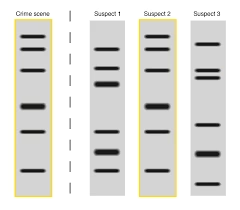
9. Reporting
Final report includes:
-
DNA profile results
-
Statistical probability
-
Interpretation
-
Expert conclusion
Applications
1. Forensic Science and Criminal Investigation
Uses:
-
Identifying suspects from blood, semen, saliva, or hair samples
-
Solving sexual assault cases
-
Linking suspects to crime scenes
-
Exonerating wrongly convicted individuals
-
Identifying repeat offenders through DNA databases
Because DNA remains stable in biological samples, even small traces can provide crucial evidence.
2. Paternity and Relationship Testing
Applications:
-
Confirming paternity or maternity
-
Resolving inheritance disputes
-
Immigration cases
-
Determining biological siblingship
Each child inherits one allele from each parent at every STR locus, making relationship analysis scientifically reliable.
3. Disaster Victim Identification (DVI)
-
Air crashes
-
Natural disasters
-
Terrorist attacks
DNA from skeletal remains, teeth, or tissues is compared with relatives’ DNA to identify victims.
4. Medical and Genetic Applications
Uses:
-
Bone marrow transplant compatibility testing
-
Detection of genetic diseases
-
Pharmacogenomics research
-
Identification of inherited disorders
DNA profiling supports personalized medicine and genetic counseling.
5. Wildlife and Conservation Biology
-
Tracking illegal poaching
-
Species identification
-
Monitoring endangered species
-
Studying genetic diversity
6. Anthropology and Evolutionary Studies
DNA fingerprinting helps:
-
Study human migration patterns
-
Trace ancestral origins
-
Analyze population genetics
Limitations
-
Risk of contamination during sample handling.
-
Requires sophisticated laboratory infrastructure.
-
Expensive equipment and reagents.
-
Requires trained personnel for interpretation.
-
Cannot distinguish identical twins.
-
Ethical and privacy concerns related to DNA databases.
-
Partial or degraded samples may give incomplete profiles.
Ethical and Legal Considerations
-
Privacy of genetic information
-
Misuse of DNA databases
-
Informed consent for DNA testing
-
Legal regulations vary across countries
Proper regulatory frameworks and ethical guidelines are essential to prevent misuse.
MCQs
1. DNA fingerprinting was discovered by:
A. Watson
B. Crick
C. Sir Alec Jeffreys
D. Mendel
Answer: C
2. DNA fingerprinting is based on:
A. Protein polymorphism
B. DNA polymorphism
C. RNA sequencing
D. Chromosome counting
Answer: B
3. The most commonly used markers in modern forensic DNA profiling are:
A. VNTR
B. STR
C. SNP only
D. mtDNA only
Answer: B
4. STR stands for:
A. Single Tandem Repeat
B. Short Tandem Repeat
C. Sequence Tandem Region
D. Simple Transcribed Repeat
Answer: B
5. STR repeat units are usually:
A. 100–200 bp
B. 50–100 bp
C. 2–6 bp
D. 500 bp
Answer: C
6. VNTRs are:
A. Coding regions
B. Non-repetitive regions
C. Repetitive non-coding sequences
D. Ribosomal RNA genes
Answer: C
7. The amplification technique used in DNA fingerprinting is:
A. ELISA
B. PCR
C. Western blot
D. Northern blot
Answer: B
8. PCR stands for:
A. Polymerase Chain Reaction
B. Protein Chain Reaction
C. Peptide Coding Reaction
D. Polymer Coding Reaction
Answer: A
9. The enzyme used in PCR is:
A. RNA polymerase
B. DNA ligase
C. Taq polymerase
D. Reverse transcriptase
Answer: C
10. DNA fragments are separated by:
A. Centrifugation
B. Chromatography
C. Electrophoresis
D. Filtration
Answer: C
11. Modern DNA profiling mainly uses:
A. Southern blotting
B. Capillary electrophoresis
C. Paper chromatography
D. SDS-PAGE
Answer: B
12. Each individual inherits STR alleles from:
A. Mother only
B. Father only
C. Both parents
D. Siblings
Answer: C
13. DNA fingerprinting cannot distinguish:
A. Siblings
B. Unrelated individuals
C. Identical twins
D. Parents
Answer: C
14. Mitochondrial DNA is inherited from:
A. Father
B. Mother
C. Both parents
D. Grandparents
Answer: B
15. Y-STR markers are useful in:
A. Maternal lineage
B. Female identification
C. Paternal lineage
D. Twin differentiation
Answer: C
16. The major application of DNA fingerprinting is:
A. Blood grouping
B. Criminal identification
C. Urine analysis
D. ECG recording
Answer: B
17. DNA fingerprinting is useful in:
A. Paternity testing
B. Diabetes diagnosis
C. Blood pressure measurement
D. Liver function test
Answer: A
18. The first step in DNA fingerprinting is:
A. PCR
B. Electrophoresis
C. Sample collection
D. Reporting
Answer: C
19. DNA quantification ensures:
A. Correct staining
B. Proper DNA concentration
C. Faster electrophoresis
D. Better gel formation
Answer: B
20. Multiplex PCR means:
A. Amplifying one gene
B. Amplifying multiple loci simultaneously
C. Sequencing DNA
D. Cutting DNA
Answer: B
21. DNA fingerprinting mainly studies:
A. Coding genes
B. Repetitive non-coding DNA
C. Ribosomal RNA
D. tRNA
Answer: B
22. The probability that two individuals share the same DNA profile is:
A. Very high
B. Moderate
C. Extremely low
D. 50%
Answer: C
23. SNP stands for:
A. Single Nucleotide Polymorphism
B. Short Nucleotide Pattern
C. Sequence Nucleic Protein
D. Small Nuclear Peptide
Answer: A
24. SNP analysis is useful in:
A. Degraded DNA samples
B. Blood grouping
C. Protein synthesis
D. RNA analysis
Answer: A
25. DNA fingerprinting is most reliable because DNA is:
A. Unstable
B. Uniform
C. Unique and stable
D. Identical in all individuals
Answer: C
26. Southern blotting was used in early analysis of:
A. STR
B. SNP
C. VNTR
D. mtDNA
Answer: C
27. Capillary electrophoresis produces:
A. Bands only
B. Peaks (electropherogram)
C. Chromosomes
D. RNA strands
Answer: B
28. DNA databases are mainly used to:
A. Store blood groups
B. Store genetic profiles
C. Store protein sequences
D. Store ECG data
Answer: B
29. DNA fingerprinting is useful in disaster victim identification because:
A. DNA survives long
B. Blood type is enough
C. Proteins are stable
D. Hair color changes
Answer: A
30. A major limitation of DNA fingerprinting is:
A. Low specificity
B. Cannot detect males
C. Risk of contamination
D. Slow PCR
Answer: C
31. Random Match Probability refers to:
A. PCR failure rate
B. Chance of random DNA match
C. Error rate in gel
D. DNA degradation rate
Answer: B
32. Forensic DNA analysis usually examines:
A. One locus
B. Two loci
C. Multiple STR loci
D. Whole genome
Answer: C
33. mtDNA is particularly useful in:
A. Fresh blood samples
B. Skeletal remains
C. Urine samples
D. Plasma proteins
Answer: B
34. DNA fingerprinting is legally accepted because it is:
A. Cheap
B. Subjective
C. Scientifically validated
D. Simple
Answer: C
35. The smallest unit of SNP variation involves:
A. Chromosome
B. Gene
C. Single base change
D. Protein
Answer: C
36. DNA fingerprinting is NOT useful in:
A. Criminal cases
B. Paternity disputes
C. Identical twin differentiation
D. Wildlife identification
Answer: C
37. The main advantage of STR markers is:
A. Large size
B. Easy PCR amplification
C. Located in coding regions
D. Stable protein coding
Answer: B
38. DNA extraction removes:
A. DNA
B. Proteins and contaminants
C. RNA only
D. Water
Answer: B
39. Forensic samples must be handled carefully to prevent:
A. Mutation
B. Amplification
C. Contamination
D. Staining
Answer: C
40. A complete DNA profile contains:
A. Protein pattern
B. Chromosome count
C. STR allele numbers
D. Blood group
Answer: C
41. DNA fingerprinting helps in:
A. Measuring glucose
B. Identifying missing persons
C. Checking ECG
D. Measuring cholesterol
Answer: B
42. The power of discrimination increases with:
A. Fewer loci
B. More loci analyzed
C. Smaller DNA
D. Less PCR cycles
Answer: B
43. DNA phenotyping predicts:
A. Blood pressure
B. Physical traits
C. Blood sugar
D. Enzyme levels
Answer: B
44. The main ethical issue in DNA fingerprinting is:
A. Cost
B. Privacy of genetic data
C. Gel preparation
D. PCR timing
Answer: B
45. Touch DNA refers to:
A. DNA from skin cells
B. DNA from saliva
C. DNA from blood
D. DNA from hair root
Answer: A
46. DNA profiling is also called:
A. Gene therapy
B. DNA typing
C. Cloning
D. Sequencing
Answer: B
47. DNA in forensic cases is often compared with:
A. Blood group
B. Database profiles
C. Urine sample
D. ECG report
Answer: B
48. The main source of variability in DNA fingerprinting is:
A. Coding genes
B. Repetitive sequences
C. Ribosomal RNA
D. Protein enzymes
Answer: B
49. PCR cycles consist of:
A. Cooling only
B. Heating only
C. Denaturation, annealing, extension
D. Washing
Answer: C
50. DNA fingerprinting is considered highly reliable because:
A. It is visual
B. It uses repetitive DNA polymorphism with statistical validation
C. It is cheap
D. It does not require expertise
Answer: B