Introduction
-
Lipids are essential biomolecules that play a critical role in cellular structure, energy storage, and signaling functions.
-
They are major components of cell membranes, myelin sheath, steroid hormones, and intracellular organelles.
-
In pathology, abnormal lipid accumulation is seen in conditions such as fatty liver (steatosis), atherosclerosis, lipid storage disorders, and certain tumors.
-
Routine histological processing with paraffin embedding often removes lipids because they are soluble in organic solvents, making special techniques necessary for their demonstration.
-
Accurate identification of lipids helps in:
-
Diagnosing metabolic and degenerative diseases
-
Identifying lipid-rich tumors
-
Studying cellular injury and inflammation
-
Research in metabolic and cardiovascular disorders
-
-
Lipid demonstration in tissues typically requires:
-
Frozen section techniques (to prevent lipid dissolution)
-
Special fat-soluble dyes
-
Advanced biochemical and microscopic methods
-
-
Different types of lipids (neutral fats, phospholipids, glycolipids, cholesterol) require specific staining and analytical approaches for accurate identification.
-
Modern histological studies combine classical staining methods with advanced techniques such as fluorescence microscopy and electron microscopy for detailed lipid analysis.
-
Therefore, lipid identification and demonstration are fundamental in both diagnostic pathology and biomedical research, enhancing our understanding of cellular function and disease mechanisms.
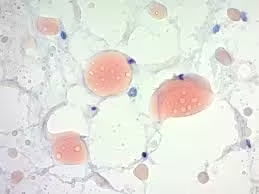
Sudan Staining
-
Reagents:
- Sudan III or Sudan IV: Dissolve 0.5 grams of dye in 100 mL of 70% alcohol (ethanol).
- Oil Red O: Dissolve 0.5 grams of Oil Red O in 100 mL of isopropanol, then dilute 6 parts Oil Red O solution with 4 parts distilled water and filter before use.
- Counterstain (optional): Harris hematoxylin stains cell nuclei blue for contrast.
-
Procedure:
-
- Preparation of Tissue Sections:
-
-
-
- Use fresh or frozen tissue sections (4–8 µm thick). Do not use paraffin-embedded tissue, as alcohol-based fixatives dissolve lipids.
-
-
-
- Fixation:
-
-
-
- Fix sections in 10% neutral-buffered formalin for 10 minutes, then rinse with distilled water.
-
-
-
- Staining:
-
-
-
- Immerse sections in the Sudan dye solution for 5–15 minutes (Oil Red O requires 10 minutes).
-
-
-
- Differentiation and Rinsing:
-
-
-
- Briefly dip the sections in 70% alcohol to remove excess dye, then rinse in distilled water.
-
-
-
- Counterstaining:
-
-
-
- Optionally stain with hematoxylin for 1–2 minutes, rinse in water and then blue the sections in a weak alkaline solution.
-
-
-
- Mounting:
-
-
-
- Mount in an aqueous medium, such as glycerin jelly, as organic solvents dissolve lipids.
-
-
-
Result:
-
- Lipid droplets and fat deposits appear bright red (Sudan III/IV) or more intense red (Oil Red O).
- Cytoplasm and other structures appear colourless or take up the counterstain (hematoxylin).
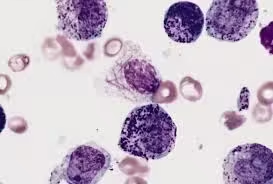
Osmium Tetroxide Staining
-
Reagents:
- Osmium Tetroxide (OsO₄): Typically prepared as a 0.5–2% solution in the buffer.
- Buffer solution: 0.1 M phosphate buffer (pH 7.4) commonly stabilises the tissue environment during fixation.
-
Procedure:
-
- Fixation:
-
-
-
- Fix fresh tissue in the OsO₄ solution (0.5–2%) for 1–2 hours at room temperature in a well-ventilated area or fume hood.
- Use small tissue pieces (1–2 mm³) for optimal penetration.
-
-
-
- Rinsing:
-
-
-
- Rinse the tissue thoroughly with phosphate buffer or distilled water to remove excess osmium.
-
-
-
- Dehydration and Embedding (optional for electron microscopy):
-
-
-
- Dehydrate tissue in graded alcohols and embed in epoxy resin if used for electron microscopy.
-
-
-
- Sectioning and Mounting:
-
-
-
- Section using a microtome, mount, and observe under a light or electron microscope for ultrastructural detail.
-
-
-
Result:
-
- Unsaturated lipids stain black as osmium binds specifically to double bonds in unsaturated fatty acids.
- Osmium-stained lipids provide contrast for electron microscopy, enhancing the visualization of lipid membranes and other lipid-rich structures.
Nile Red Staining
-
Reagents:
- Nile Red: Dissolve Nile Red powder in acetone to prepare a 1 mg/mL stock solution.
- Working Solution: Dilute to 1–10 µg/mL in distilled water or PBS immediately before use.
- Counterstain (optional): DAPI or other nuclear stains for fluorescence.
-
Procedure:
-
- Preparation of Tissue or Cells:
-
-
-
- Use either live or fixed tissue sections or cultured cells.
-
-
-
- Application of Dye:
-
-
-
- Apply Nile Red working solution to the sample and incubate for 10–15 minutes at room temperature, protected from light.
-
-
-
- Washing:
-
-
-
- Rinse gently with PBS or distilled water to remove excess dye.
-
-
-
- Imaging:
-
-
-
- Visualize using a fluorescence microscope with filters for Nile Red excitation (usually 488 nm) and emission.
-
-
-
Result:
-
- Neutral lipids in lipid droplets fluoresce yellow-gold, while phospholipids show red fluorescence.
- The distinct colours enable differentiation between lipid classes.
Fluorescent Lipid Dyes Staining (e.g., BODIPY)
-
Reagents:
- BODIPY Lipid Dyes: Available in various conjugates, typically prepared as 1–10 µM in DMSO or ethanol.
- PBS or suitable buffer: Used for washing.
-
Procedure:
-
- Preparation of Sample:
-
-
-
- Fix tissue sections or cultured cells if necessary. BODIPY can also be used in live cells.
-
-
-
- Application of Dye:
-
-
-
- Incubate the sample with BODIPY solution for 5–15 minutes, protected from light.
-
-
-
- Washing:
-
-
-
- Rinse with PBS to remove unbound dye.
-
-
-
- Microscopy:
-
-
-
- Visualize using a fluorescence microscope with specific filters for BODIPY (typically ~500 nm excitation and 510–550 nm emission).
-
-
-
Result:
-
- Lipid droplets and membrane structures stain bright green or yellow-green.
- BODIPY’s photostability makes it ideal for time-lapse imaging, allowing for live tracking of lipid dynamics.
Enzyme Histochemistry for Lipid Detection
-
Reagents:
- Enzymes: Lipase (for triglycerides) and phospholipase (for phospholipids) are commonly used.
- Substrate or Chromogen: Fast Blue or Fast Red can produce a coloured reaction product.
- Buffer: Specific for each enzyme, such as Tris buffer for phospholipase.
-
Procedure:
-
- Enzyme Incubation:
-
-
-
- Incubate tissue sections with enzyme solution (e.g., lipase in 0.1 M Tris buffer, pH 7.4) at 37°C for 30–120 minutes.
-
-
-
- Substrate Reaction:
-
-
-
- After enzyme treatment, apply the substrate solution and incubate. The substrate reacts with the enzymatic degradation products to form a coloured product.
-
-
-
- Counterstaining (optional):
-
-
-
- Counterstain with hematoxylin to enhance contrast.
-
-
-
- Mounting:
-
-
-
- Mount sections in an aqueous medium for microscopic examination.
-
-
-
Result:
-
- Areas containing the target lipid type will develop a colour, indicating enzyme specificity.
- For example, triglycerides produce a coloured product with lipase staining, while phospholipids stain with phospholipase.
Applications:
- Disease Diagnosis: Abnormal lipid accumulation can indicate metabolic diseases, atherosclerosis, or fatty liver disease.
- Research on Lipid Metabolism: Identifying and localizing lipids helps understand lipid metabolism and cell signalling pathways.
- Tissue Engineering: In regenerative medicine, understanding lipid presence can influence scaffold development for cell growth.