- Quantifying bacterial populations is essential for understanding their roles in various environments, including natural habitats, clinical settings, food safety, and biotechnological applications.
- Two main types of bacterial counts are commonly performed: total bacterial count and viable bacterial count.
- These methods provide insights into the overall microbial load and the proportion of living or potentially active cells within a sample.
Total Bacterial Count
- Total bacterial count estimates the overall bacterial population within a sample, regardless of whether the cells are alive or dead.
- This measure is essential in contexts where bacterial biomass or general population dynamics are of interest, such as environmental studies, microbiome research, or quality control in industrial applications.
- Methods for Total Bacterial Count
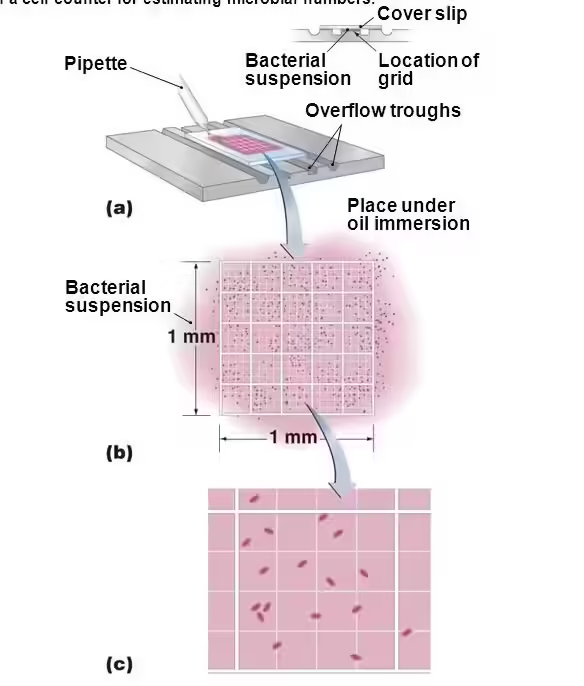
1.1 Direct Microscopic Count (DMC)
The direct microscopic count is one of the simplest and most direct methods to measure the total bacterial count.
- Procedure:
- First, a small volume of the bacterial sample is placed on a specialized counting slide, often a Petroff-Hausser or Neubauer chamber, which contains a grid marked with squares of known area and depth.
- A known volume of the bacterial suspension is applied to the grid, which helps estimate the concentration of bacteria in the entire sample by counting the cells within each square.
- After loading the sample, the slide is placed under a microscope, typically at 400x or 1000x magnification, and bacterial cells within several squares are counted.
- Applications:
- DMC is widely used in dairy microbiology to assess bacterial loads in milk and in environmental microbiology to evaluate microbial biomass in water or soil.
- This method is also valuable in studying microbial ecology and population dynamics.
- Limitations and Variations:
- Live/Dead Staining: DMC can be combined with fluorescent stains to distinguish between live and dead cells. For example, the SYTO9 and propidium iodide stains help to selectively stain live and dead bacteria, making it easier to distinguish viable cells.
- Limitations: Without staining, this method cannot differentiate between viable and dead cells. It is also prone to human error, especially in samples with very high or low bacterial densities.
1.2 Flow Cytometry
Flow cytometry is an advanced, automated method that can analyze thousands of cells per second.
- Procedure:
- The bacterial suspension is loaded into a flow cytometer, where cells pass through a laser beam one at a time. As each cell passes through, it scatters light and emits fluorescence, which can be detected and quantified.
- Bacteria can be pre-stained with DNA-binding fluorescent dyes, such as DAPI or SYBR Green to enhance cell detection.
- Light scattering patterns help determine cell size and structure, while fluorescence intensity helps quantify total cell numbers.
- Applications:
- Environmental microbiology: Flow cytometry analyzes bacterial populations in marine and freshwater environments.
- Medical microbiology: Employed in clinical laboratories to quickly assess bacterial presence in patient samples.
- Industrial applications: Used in bioreactors and fermentation processes to monitor microbial load in real-time.
- Limitations and Variations:
- Cost: Flow cytometers are expensive, making them less accessible for routine analysis.
- Viability Distinction: Like DMC, this method requires specific staining to differentiate between live and dead cells, as standard flow cytometry does not distinguish cell viability.
1.3 Turbidity Measurement
Turbidity is a simple, rapid method often used to monitor bacterial growth in real-time by measuring a culture’s optical density (OD).
- Procedure:
- A spectrophotometer measures the amount of light passing through the bacterial suspension at a specific wavelength, usually OD600 since bacterial cells absorb at this wavelength.
- The OD value correlates with cell density. A standard curve, generated by plotting OD values against known bacterial concentrations, can then estimate the total cell count in unknown samples.
- Applications:
- Biotechnology and research: Commonly used to monitor bacterial growth in liquid cultures.
- Food and beverage industries: Employed to assess the bacterial content in liquid samples like broths, juices, and other non-viscous products.
- Limitations and Variations:
- Non-specificity: Turbidity measures live and dead cells, making it unsuitable for viability assessments without additional testing.
- Limitations at Low Densities: OD readings may not be sensitive enough to detect bacterial presence at low cell concentrations.
Viable Bacterial Count
- The viable bacterial count quantifies the number of living, actively dividing cells, also known as colony-forming units (CFUs).
- This count is crucial for applications where cell viability is essential, such as clinical diagnostics, food safety, and quality assurance.
- Methods for Viable Bacterial Count
1.1 Plate Count Method
The plate count method, or colony-forming unit (CFU), is one of the most widely used techniques to determine viable bacterial counts.
- Procedure:
- A bacterial sample is serially diluted to ensure countable numbers on agar plates.
- A known volume of each dilution is spread onto the surface of an agar plate (spread plate method) or mixed with molten agar before it solidifies (pour plate method).
- After incubation, colonies are counted, and CFU per ml or gram is calculated by multiplying the counted colonies by the dilution factor.
- Applications:
- Food microbiology: Used to detect and quantify foodborne pathogens and spoilage organisms.
- Pharmaceutical and cosmetic industries: Helps ensure product sterility and bacterial quality control.
- Water testing: Common in public health labs to assess drinking water quality.
- Limitations and Variations:
- Selective Media: Different media types can be used to selectively grow certain bacteria, which can aid in detecting specific groups like coliforms or pathogens.
- Time-Consuming: Results may take 1-2 days, and only culturable bacteria that grow under specific conditions will be detected.
- Spread vs. Pour Plates: Spread plates are useful for aerobic bacteria, while pour plates are often employed for aerobic and anaerobic bacteria.
1.2 Most Probable Number (MPN) Method
The MPN method is a statistical estimation technique commonly used for samples with low bacterial counts, such as drinking water.
- Procedure:
- A series of dilutions are inoculated into multiple tubes containing a liquid growth medium. After incubation, each tube is checked for bacterial growth (often indicated by a color change or turbidity).
- Statistical tables are used to estimate the original sample’s viable bacterial concentration based on the number of positive (turbid) tubes at each dilution.
- Applications:
- Water quality: Frequently used in detecting coliform bacteria and Escherichia coli in water, important indicators of fecal contamination.
- Soil microbiology: Useful for enumerating soil bacteria that may not grow on solid media.
- Limitations and Variations:
- Accuracy: MPN is less precise than plate counts and relies on probabilistic estimation.
- Labor Intensive: Requires multiple replicates for each dilution level and takes time to interpret.
1.3 Membrane Filtration
Membrane filtration is especially useful for water samples and allows for the concentration of bacteria from large volumes.
- Procedure:
- A known volume of water is passed through a membrane filter with pores small enough (usually 0.45 µm) to capture bacteria.
- The filter is then placed on an agar plate and incubated to allow for colony formation, which can then be counted.
- Applications:
- Environmental and public health: Common in assessing water quality and monitoring bacterial contamination in drinking water, rivers, and oceans.
- Food testing: Suitable for liquid samples like juices and broths.
- Limitations and Variations:
- Limitations for Particle-Rich Samples: Filters may clog if the sample has high particle content.
- Specific Media: Different agar types can target specific bacterial groups, such as coliforms, which are essential indicators in water testing.
Advanced Methods and Viability Distinction Techniques
Fluorescent Staining with Viability Dyes:
- Modern methods utilize fluorescent stains like SYTO9 and propidium iodide, where SYTO9 stains live cells green, and propidium iodide stains dead cells red. Combined with microscopy or flow cytometry, this approach allows for quick differentiation between viable and non-viable cells.
Quantitative PCR with Propidium Monoazide (PMA):
- PMA binds to DNA in dead cells, preventing amplification during PCR. By comparing DNA concentrations with and without PMA treatment, it’s possible to estimate the number of live cells, providing a robust measure of viable bacteria.
Practical Applications in Various Fields
Food Safety:
- Total and viable counts are vital for assessing bacterial load in food products, ensuring compliance with food safety standards, and preventing spoilage and foodborne diseases.
Water Quality Testing:
- Viable bacterial counts are essential in monitoring public water supplies for pathogens, while total counts provide an overview of the microbial load and potential biofilm formation risks.
Environmental Microbiology:
- Total bacterial counts help assess microbial biomass, whereas viable counts focus on metabolically active communities, which is crucial for understanding microbial ecosystem functions.
Clinical Microbiology:
- Viable counts provide important information about the severity of infections and are used in diagnostic labs to guide treatment and track infection progression.